| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,551,170 – 7,551,263 |
| Length | 93 |
| Max. P | 0.900676 |

| Location | 7,551,170 – 7,551,263 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 63.09 |
| Shannon entropy | 0.75104 |
| G+C content | 0.48558 |
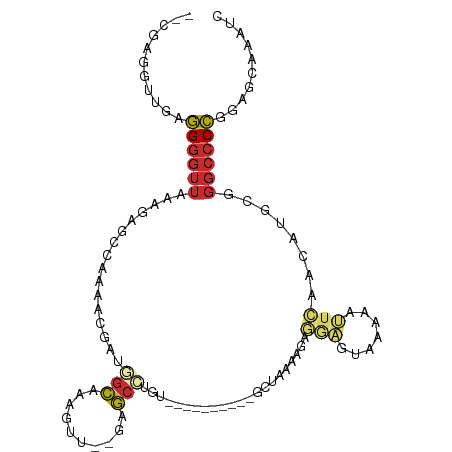
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -7.93 |
| Energy contribution | -8.36 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900676 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
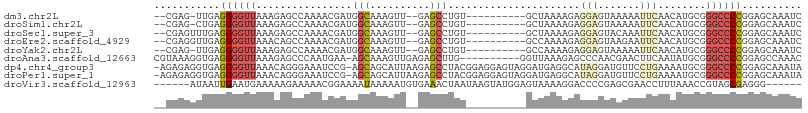
>dm3.chr2L 7551170 93 + 23011544 --CGAG-UUGAGGGGUUAAAGAGCCAAAACGAUGGCAAAGUU--GAGCCUGU----------GCUAAAAGAGGAGUAAAAAUUCAACAUGCGGGCCCCGGAGCAAAUC --...(-((..((((((.....((((......))))...(((--(((....(----------(((........))))....)))))).....))))))..)))..... ( -27.20, z-score = -2.28, R) >droSim1.chr2L 7346798 93 + 22036055 --CGAG-CUGAGGGGUUAAAGAGCCAAAACGAUGGCAAAGUU--GAGCCUGU----------GCUAAAAGAGGAGUAAAAAUUCAACAUGCGGGCCCCGGAGCAAAUC --...(-((..((((((.....((((......))))...(((--(((....(----------(((........))))....)))))).....))))))..)))..... ( -29.60, z-score = -2.67, R) >droSec1.super_3 3064816 94 + 7220098 --CGAGUUUGAGGGGUUAAAGAGCCAAAACGAUGGCAAAGUU--GAGCCUGU----------GCUAAAAGAGGAGUACAAAUUCAACAUGCGGGCCCCGGAGCAAAUC --...((((..((((((.....((((......))))...(((--(((..(((----------(((........))))))..)))))).....)))))).))))..... ( -31.80, z-score = -3.36, R) >droEre2.scaffold_4929 16469599 94 + 26641161 --CGAGGUUGAGGGGUUAAACAGCCAAAACGAUGGCAAAGUU--GAGCCUGU----------GCCAAAAGAGGAGUAAGAAUUCAACAUGCGGGCCCCGGAGCAAAUC --....(((..((((((...((((((......))))...(((--(((.((..----------.((......))....))..)))))).))..))))))..)))..... ( -26.40, z-score = -1.26, R) >droYak2.chr2L 16978692 93 - 22324452 --CGAG-UUGAGGGGUUAAAGAGCCAAAACGAUGGCAAAGUU--GAGCCUGU----------GCCAAAAGAGGAGUAAAAAUUCAACAUGCGGGCCCCGGAGCAAAUC --...(-((..((((((.....((((......))))...(((--((((((.(----------......).)))........)))))).....))))))..)))..... ( -25.80, z-score = -1.62, R) >droAna3.scaffold_12663 1473 97 + 19816 CGUAAAGGUGAGGGGUUAAAGAGCCCAAUGAA-AGCAAAGUUGAGAGCUUG----------GGUUAAAGAGCCCAACGAACUUCAAUAUGCGGGCCCCGGAGCCAAAC ......(((..((((((...(....)......-.(((..((((((..((((----------((((....))))))).)...)))))).))).))))))...))).... ( -28.60, z-score = -1.59, R) >dp4.chr4_group3 1423414 106 + 11692001 -AGAGAGGUGAGGGGUUAAACAGGGAAAUCCG-AGCAGCAUUAAGAGCCUACGGAGGAGUAGGAUGAGGCAUAGGAUGUUCCUGAAAAUGCGGGCCCCGGAGCAAAUA -......((..((((((...((((((.((((.-.((..(((......(((((......))))))))..))...)))).))))))........))))))...))..... ( -33.90, z-score = -2.74, R) >droPer1.super_1 2892047 106 + 10282868 -AGAGAGGUGAGGGGUUAAACAGGGAAAUCCG-AGCAGCAUUAAGAGCCUACGGAGGAGUAGGAUGAGGCAUAGGAUGUUCCUGAAAAUGCGGGCCCCGGAGCAAAUA -......((..((((((...((((((.((((.-.((..(((......(((((......))))))))..))...)))).))))))........))))))...))..... ( -33.90, z-score = -2.74, R) >droVir3.scaffold_12963 11002559 96 + 20206255 ------AUAAUUGAAUGAAAAAGAAAAACGGAAAAUAAAAAUGUGAAACUAAUAAGUAUGGAGUAAAAGGACCCCGAGCGAACCUUUAAACCGUAGCGAGGG------ ------.....................((((..........(((.......))).((.(((.((......)).))).))...........))))........------ ( -8.40, z-score = 1.11, R) >consensus __CGAGGUUGAGGGGUUAAAGAGCCAAAACGAUGGCAAAGUU__GAGCCUGU__________GCUAAAAGAGGAGUAAAAAUUCAACAUGCGGGCCCCGGAGCAAAUC ...........((((((................(((..........)))......................(((.......)))........)))))).......... ( -7.93 = -8.36 + 0.42)
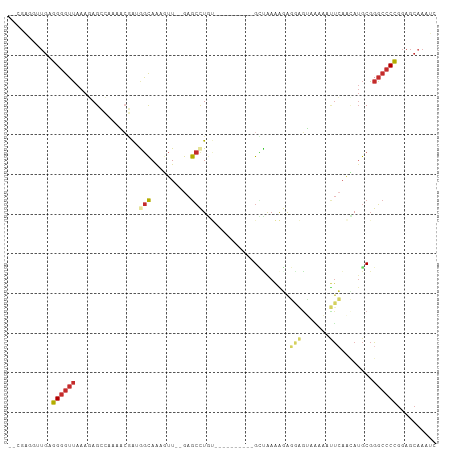
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:54 2011