| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,476,417 – 11,476,508 |
| Length | 91 |
| Max. P | 0.999402 |

| Location | 11,476,417 – 11,476,508 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 66.89 |
| Shannon entropy | 0.64676 |
| G+C content | 0.54194 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -14.14 |
| Energy contribution | -13.48 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
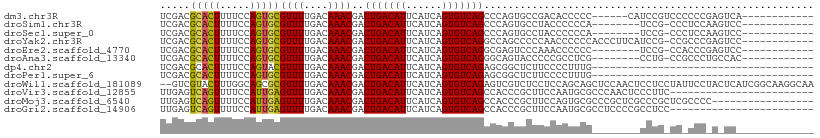
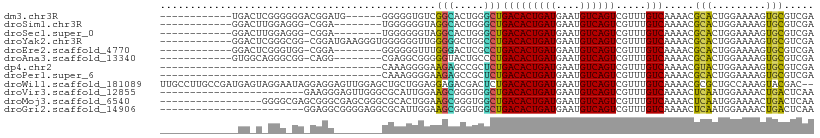
>dm3.chr3R 11476417 91 + 27905053 UCGACGCACUUUUCCAGUGCGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGCCCAGUGCCGACACCCCC------CAUCCGUCCCCCCGAGUCA------------ ..((((((((.....)))))))).((((......(((((((......)))))))....(((....)))....------...............))))------------ ( -23.00, z-score = -3.03, R) >droSim1.chr3R 17509274 88 - 27517382 UCGACGCACUUUUCCAGUGCGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGCCCAGUGCCUACCCCCCA--------UCCG-CCCUCCAAGUCC------------ ..((((((((.....))))))))..(((......(((((((......)))))))...((.((..........--------...)-).))....))).------------ ( -20.22, z-score = -3.08, R) >droSec1.super_0 10647109 88 - 21120651 UCGACGCACUUUUCCAGUGCGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGCCCAGUGCCUACCCCCCA--------UCCG-CCCUCCAAGUCC------------ ..((((((((.....))))))))..(((......(((((((......)))))))...((.((..........--------...)-).))....))).------------ ( -20.22, z-score = -3.08, R) >droYak2.chr3R 13870571 96 - 28832112 UCGACGCACUUUUCCAGUGCGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGGCCAGCCCCCAACCCCCCACCCUUCAUCCG-CCGCCCGAGUCC------------ ..((((((((.....)))))))).........(((((((((......))))).(((..((.......................)-).)))..)))).------------ ( -21.30, z-score = -2.35, R) >droEre2.scaffold_4770 10991526 88 + 17746568 UCGACGCACUUUUCCAGUGCGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGGCGAGUCCCAAACCCCCC--------UCCG-CCACCCGAGUCC------------ ..((((((((.....)))))))).........(((((((((......))))).((((((............)--------).))-)).....)))).------------ ( -24.90, z-score = -3.17, R) >droAna3.scaffold_13340 16084639 88 + 23697760 UCGACGCACUUUUCCAGUGCGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGGGCAGUACCCCCGCCUCG--------CCUG-CCGCCCUGCCAC------------ ..((((((((.....))))))))...........(((((((......)))))))(((((..(....((....--------...)-).)..)))))..------------ ( -27.00, z-score = -2.33, R) >dp4.chr2 30480601 72 + 30794189 UCGACGCACUUUUCCAGUACGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGAGCGGCUCUUCCCCUUUG------------------------------------- ..((.(((((.....))).(((((((((.....(((((......))))))))))))))))))..........------------------------------------- ( -16.90, z-score = -1.71, R) >droPer1.super_6 5877144 72 + 6141320 UCGACGCACUUUUCCAGUGCGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGAGCGGCUCUUCCCCUUUG------------------------------------- ..((((((((.....))))))))........((.(((((((......)))))))..))..............------------------------------------- ( -20.20, z-score = -2.29, R) >droWil1.scaffold_181089 9915894 107 - 12369635 --GUCGUACUUUGGCAGCGCGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGAGUCGUCUCCUCCAGCAGCUCCAACUCCUCCUAUUCCUACUCAUCGGCAAGGCAA --(((.....((((.(((((.....((...(((((((((((......)))))..)))))).)).....)).)))))))...........((........))...))).. ( -23.40, z-score = -0.34, R) >droVir3.scaffold_12855 5090492 85 + 10161210 UUGAGUCAGUUUUCCAUUGAGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGCCACCCGCUUCCAAUGCGCCCAACUCCCUUC------------------------ ....(((((..(((....)))..)))))....(.(((((((......))))))).)...(((.......))).............------------------------ ( -18.50, z-score = -2.25, R) >droMoj3.scaffold_6540 22367396 92 + 34148556 UUGAGUCAGUUUUCCAUUGAGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGCCACCCGCUUCCAGUGCGCCCGCUCGCCCGCUCGCCCC----------------- ....(((((..(((....)))..)))))....(.(((((((......))))))).)....((....((((.((......)).)))).))...----------------- ( -20.80, z-score = -1.18, R) >droGri2.scaffold_14906 4752799 85 - 14172833 UUGAGUCAGUUUUCCAUUGAGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGCCACCCGCUUCCAAUGCGCCUCCCCGCCUCC------------------------ ....(((((..(((....)))..)))))....(.(((((((......))))))).)...(((.......))).............------------------------ ( -18.50, z-score = -2.10, R) >consensus UCGACGCACUUUUCCAGUGCGUUUUGACAAACGACUGACAUUCAUCAGUGUCAGCCCAGCGCCUCCCCCCCG________CCCG_CC_CCC_A_UC_____________ .....(((((.....)))))((((....))))..(((((((......)))))))....................................................... (-14.14 = -13.48 + -0.65)
| Location | 11,476,417 – 11,476,508 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 66.89 |
| Shannon entropy | 0.64676 |
| G+C content | 0.54194 |
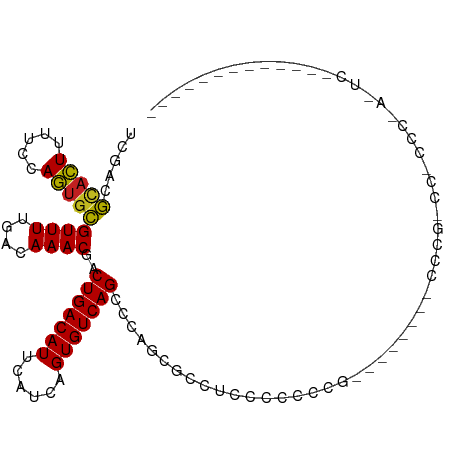
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -9.82 |
| Energy contribution | -8.81 |
| Covariance contribution | -1.01 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.678013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
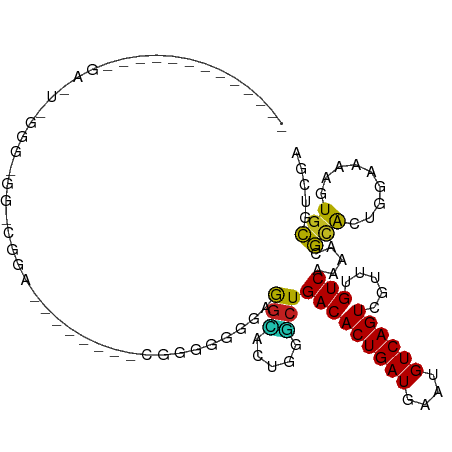
>dm3.chr3R 11476417 91 - 27905053 ------------UGACUCGGGGGGACGGAUG------GGGGGUGUCGGCACUGGGCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGCACUGGAAAAGUGCGUCGA ------------....(((....((((((((------(..(((((((((.....)))))))))(((....)))..)))))))))...(((((((.....)))))))))) ( -35.90, z-score = -3.20, R) >droSim1.chr3R 17509274 88 + 27517382 ------------GGACUUGGAGGG-CGGA--------UGGGGGGUAGGCACUGGGCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGCACUGGAAAAGUGCGUCGA ------------.(((......((-((((--------((....((.(((.....))).))((((((....)))))))))))))).....(((((.....)))))))).. ( -28.00, z-score = -1.55, R) >droSec1.super_0 10647109 88 + 21120651 ------------GGACUUGGAGGG-CGGA--------UGGGGGGUAGGCACUGGGCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGCACUGGAAAAGUGCGUCGA ------------.(((......((-((((--------((....((.(((.....))).))((((((....)))))))))))))).....(((((.....)))))))).. ( -28.00, z-score = -1.55, R) >droYak2.chr3R 13870571 96 + 28832112 ------------GGACUCGGGCGG-CGGAUGAAGGGUGGGGGGUUGGGGGCUGGCCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGCACUGGAAAAGUGCGUCGA ------------....(((...((-(((((((..((((..((((..(...)..))))..))))(((....)))..)))))))))...(((((((.....)))))))))) ( -35.40, z-score = -2.20, R) >droEre2.scaffold_4770 10991526 88 - 17746568 ------------GGACUCGGGUGG-CGGA--------GGGGGGUUUGGGACUCGCCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGCACUGGAAAAGUGCGUCGA ------------.(((..(..(((-((.(--------((.(((((...))))).)))(((((((((....)))))).))))))))...)(((((.....)))))))).. ( -30.90, z-score = -1.33, R) >droAna3.scaffold_13340 16084639 88 - 23697760 ------------GUGGCAGGGCGG-CAGG--------CGAGGCGGGGGUACUGCCCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGCACUGGAAAAGUGCGUCGA ------------.((((.....((-((((--------((((((((.....)))))((((((........))))))))))))))).....(((((.....))))))))). ( -37.60, z-score = -2.68, R) >dp4.chr2 30480601 72 - 30794189 -------------------------------------CAAAGGGGAAGAGCCGCUCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGUACUGGAAAAGUGCGUCGA -------------------------------------((.((.((.....)).)).))....(((..((((.....))))..)))..(((((((.....)))))))... ( -19.40, z-score = -0.91, R) >droPer1.super_6 5877144 72 - 6141320 -------------------------------------CAAAGGGGAAGAGCCGCUCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGCACUGGAAAAGUGCGUCGA -------------------------------------((.((.((.....)).)).))....(((..((((.....))))..)))..(((((((.....)))))))... ( -21.40, z-score = -1.23, R) >droWil1.scaffold_181089 9915894 107 + 12369635 UUGCCUUGCCGAUGAGUAGGAAUAGGAGGAGUUGGAGCUGCUGGAGGAGACGACUCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGCGCUGCCAAAGUACGAC-- ...(((..((........))...))).(...(((((((.((....(....)(((...(((((((((....)))))).))).))).....))))).))))....)...-- ( -28.80, z-score = 0.04, R) >droVir3.scaffold_12855 5090492 85 - 10161210 ------------------------GAAGGGAGUUGGGCGCAUUGGAAGCGGGUGGCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACUCAAUGGAAAACUGACUCAA ------------------------.....((((..(.(.((((((..(((..(((((((((........)))))))))..)))......)))))))....)..)))).. ( -28.10, z-score = -2.75, R) >droMoj3.scaffold_6540 22367396 92 - 34148556 -----------------GGGGCGAGCGGGCGAGCGGGCGCACUGGAAGCGGGUGGCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACUCAAUGGAAAACUGACUCAA -----------------(((..(((..((((((((..(((.(((....))))))(((((((........)))))))))))))))....)))...(.....)...))).. ( -27.70, z-score = -0.40, R) >droGri2.scaffold_14906 4752799 85 + 14172833 ------------------------GGAGGCGGGGAGGCGCAUUGGAAGCGGGUGGCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACUCAAUGGAAAACUGACUCAA ------------------------.(((.(((.....(.((((((..(((..(((((((((........)))))))))..)))......)))))))....))).))).. ( -25.90, z-score = -1.60, R) >consensus _____________GA_U_GGG_GG_CGGA________CGGGGGGGAGGCACUGGGCUGACACUGAUGAAUGUCAGUCGUUUGUCAAAACGCACUGGAAAAGUGCGUCGA ..............................................(((.....)))(((((((((....)))))).....))).....(((.........)))..... ( -9.82 = -8.81 + -1.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:02 2011