| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,468,521 – 11,468,642 |
| Length | 121 |
| Max. P | 0.859295 |

| Location | 11,468,521 – 11,468,642 |
|---|---|
| Length | 121 |
| Sequences | 7 |
| Columns | 142 |
| Reading direction | forward |
| Mean pairwise identity | 69.43 |
| Shannon entropy | 0.56971 |
| G+C content | 0.57992 |
| Mean single sequence MFE | -43.03 |
| Consensus MFE | -21.67 |
| Energy contribution | -23.16 |
| Covariance contribution | 1.49 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11468521 121 + 27905053 CCUUCAAGUGCUCAAUUGUGCAAACUCCUUGAAUGGCCAGUAGAU-------------GGGGAGACCCGCCUCUGCUUGGCCAGCUGCUCCGCCACUGACUCCCGUGGCAGAGUCUCCGAUCCUAAGCUCGAGC-------- ..(((((((((.(....).))).....))))))((((((((((((-------------(((....))))..))))).))))))(((((((.(((((........))))).))))...(((........))))))-------- ( -47.00, z-score = -2.50, R) >droSim1.chr3R 17501341 121 - 27517382 CCUUCAAGUGCUCAAUUGUGCAAACUCCUUGAAUGGCCGGUAGAU-------------GGGGAGACCCGCCUCUGCUUGGCCAGCUGCUCCGCCACUGACUCCCGUGGCAGAGUCGGCGAUCCUAAGCUCGAGC-------- .......(..(......)..)......(((((.((((((((((((-------------(((....))))..))))).))))))(((((((.(((((........))))).))).))))..........))))).-------- ( -50.80, z-score = -2.58, R) >droSec1.super_0 10639187 121 - 21120651 CCUUCAAGUGCUCAAUUGUGCAAACUCCUUGAAUGGCCAGUAGAU-------------GGGGAGACCCGCCUCUGCUUGGCCAGCUGCUCCGCCACUAACUCCCGUGGCAGAGACUCCGAUCCUAAGCUCGAGC-------- ..(((((((((.(....).))).....))))))((((((((((((-------------(((....))))..))))).))))))...((((.(((((........))))).(((.((.........)))))))))-------- ( -46.30, z-score = -2.76, R) >droYak2.chr3R 13862550 132 - 28832112 --UUCAAGUGCUCAAUUGUGCCAACUCCUUGAAUGGCCAGAAGAUGGACCAGGAGAUGGGGCAGACCCGCCUCUGCUUGGCCAGCUGCUCUGCCACCGACUCCCGUGGCGGAGUCUCCGAUCUUAAGCUUAAAC-------- --.......(((...(((.((((..........)))))))(((((((.(((((....(((((......)))))..)))))))....((((((((((.(....).)))))))))).....))))).)))......-------- ( -45.20, z-score = -0.68, R) >droEre2.scaffold_4770 10983330 121 + 17746568 CGUUCAAGUGCUCAAUUGUGCAAACUCCCUGAAUGGCCAAA-------------GAUGGGGCAGACCCGCCUCUGCUUGGCCAGCUGCUUUGCCACUGACUCCCGCGGCACAGUCUCCGAUCCUGAGCUCGAGC-------- .((((....(((((((((...............(((((((.-------------...(((((......)))))...)))))))((((...((((.(........).))))))))...))))..)))))..))))-------- ( -37.10, z-score = 0.47, R) >droAna3.scaffold_13340 16077474 96 + 23697760 CAUUCAAGUGUUCAAUUGUGCAAACUCCUUGAAUGGCA-----------------------------CGCCUCUACUUGGUCAGCUGCUCCGCCAGUGGCUCCUGUGGCACAGUAUUUGAUCGAG----------------- (((((((((((........))).....))))))))...-----------------------------........(((((((((.((((..((((..(....)..))))..)))).)))))))))----------------- ( -27.60, z-score = -0.82, R) >droMoj3.scaffold_6540 22357380 128 + 34148556 --UUUAAGUGUGCAAUUGUUCAAGCCUCUUGCAUGGCUCGAGUUGU------------GGACGGCUGUGCCCCUGGCUGUCCCCUGCGACUGCCGCUCUCCGCUCUGGCAGACGAGUGCCUCUACAGCUCCUGUGCCCGGGC --....((((.(((.....((.((((........)))).))(((((------------((((((((........))))))))...))))))))))))....((((.((((.(.((((.........)))).).)))).)))) ( -47.20, z-score = -0.82, R) >consensus CCUUCAAGUGCUCAAUUGUGCAAACUCCUUGAAUGGCCAGAAGAU_____________GGGCAGACCCGCCUCUGCUUGGCCAGCUGCUCCGCCACUGACUCCCGUGGCAGAGUCUCCGAUCCUAAGCUCGAGC________ ..(((((((((.(....).))).....))))))(((((((..................(((....)))((....)))))))))...((((.(((((........))))).))))............................ (-21.67 = -23.16 + 1.49)

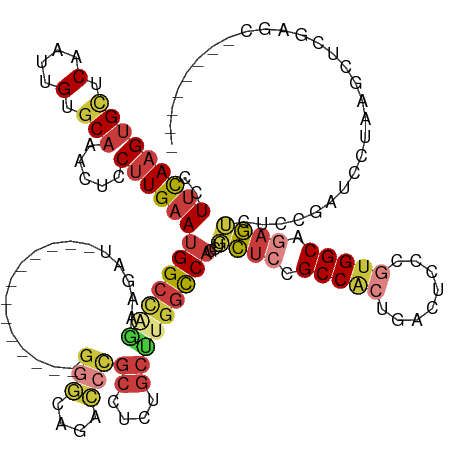

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:01 2011