
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,454,100 – 11,454,194 |
| Length | 94 |
| Max. P | 0.620433 |

| Location | 11,454,100 – 11,454,194 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 84.58 |
| Shannon entropy | 0.31050 |
| G+C content | 0.40875 |
| Mean single sequence MFE | -21.29 |
| Consensus MFE | -14.20 |
| Energy contribution | -13.74 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
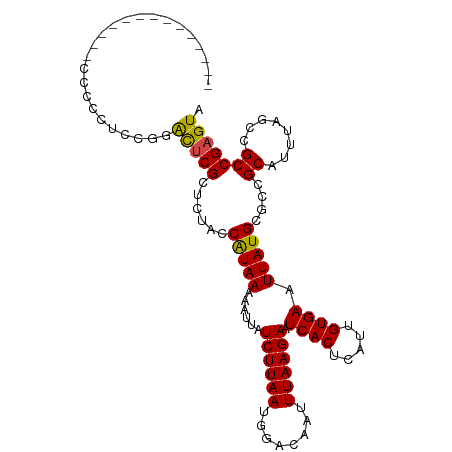
>dm3.chr3R 11454100 94 - 27905053 --------------UCCCCUACGGACUCGCUCUACCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGCCGCAUUUAGCCGCCGAGUA --------------....(....)(((((......(((((......((((((........)))))).((((.....)))).))))).((.((.....)).))))))). ( -18.80, z-score = -1.63, R) >droSim1.chr3R 17485428 94 + 27517382 --------------UCCCCUACGGACUCGCUCUACCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGCCGCAUUUAGCCGCCGAGUA --------------....(....)(((((......(((((......((((((........)))))).((((.....)))).))))).((.((.....)).))))))). ( -18.80, z-score = -1.63, R) >droSec1.super_0 10623411 94 + 21120651 --------------UCCCCUGCGGACUCGCUCUACCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGCCGCAUUUAGCCGCCGAGUA --------------..........(((((......(((((......((((((........)))))).((((.....)))).))))).((.((.....)).))))))). ( -18.70, z-score = -1.08, R) >droYak2.chr3R 13847488 95 + 28832112 -------------UCCCCCACCGGACUCGCUCUACCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGCUGCAUUUACCCGCCGAGUA -------------...........(((((......(((((......((((((........)))))).((((.....)))).)))))....((........))))))). ( -15.40, z-score = -1.07, R) >droEre2.scaffold_4770 10968871 95 - 17746568 -------------UCCCCCAACGCACGCGCUCCACCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGCUGCAUUUACCCGCCGAGUA -------------.........(((.((((................((((((........)))))).((((.....)))).....)))))))................ ( -19.00, z-score = -2.39, R) >droAna3.scaffold_13340 16065162 95 - 23697760 -------------UCCCCUUUUGGACUCGCUCUACCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGCUGCAUUUAGCCGCCGAGUA -------------...((....))(((((......(((((......((((((........)))))).((((.....)))).))))).((.((.....)).))))))). ( -19.80, z-score = -1.99, R) >dp4.chr2 30454634 94 - 30794189 --------------UCUCCUUCAGGCUCGUUCUACCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGGUGCAUUUAGCUGCCGAGUA --------------..........((((.......(((((......((((((........)))))).((((.....)))).))))).(((((.....)).))))))). ( -19.20, z-score = -1.17, R) >droPer1.super_6 5850998 94 - 6141320 --------------UCUCCUUCUGGCUCGUUCUACCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGGUGCAUUUAGCUGCCGAGUA --------------..........((((.......(((((......((((((........)))))).((((.....)))).))))).(((((.....)).))))))). ( -19.20, z-score = -1.29, R) >droWil1.scaffold_181089 9895241 108 + 12369635 UCUCCUUCUGGCCACGAUGCCGUUGCUCGCUCUAGCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGGUGCAUUUAGUUGCCGAGUA .........(((......)))..((((((.....((((((......((((((........)))))).((((.....)))).))))))...(((......))))))))) ( -26.10, z-score = -1.27, R) >droVir3.scaffold_12855 5062186 90 - 10161210 ------------------UCCCCUGUUCGCUCUGGCGUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGGCGCAUUUAGCUGCCGAGUA ------------------..........((((..((((((......((((((........)))))).((((.....)))).))))))(((((.....)).))))))). ( -24.50, z-score = -2.27, R) >droMoj3.scaffold_6540 22340027 101 - 34148556 -------GGCAGCUCCCCUGUGCUGUGCGCUCUGGCGUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGGCGCAUUUAGCUGCCGAGUA -------(((((((....((((((((((((....))))........((((((........)))))).((((.....)))).....))))))))...)))))))..... ( -37.10, z-score = -3.71, R) >droGri2.scaffold_14906 4725058 90 + 14172833 ------------------UCCCCUGUUCGUUCUGGCGUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGGCGCAUUUAGUUGCCGAGUA ------------------.......(((......((((((......((((((........)))))).((((.....)))).))))))((((........))))))).. ( -18.90, z-score = -0.70, R) >consensus ______________CCCCCUCCGGACUCGCUCUACCAUAAAAAUUAUCUUAAUGGACAAUUUAAGAAUCACUCAUUGUGAAUUAUGCGCCGCAUUUAGCCGCCGAGUA ........................(((((......(((((......((((((........)))))).((((.....)))).)))))....((........))))))). (-14.20 = -13.74 + -0.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:18:00 2011