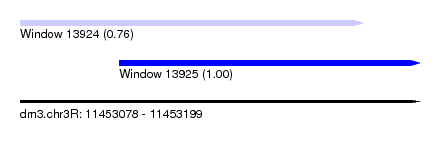
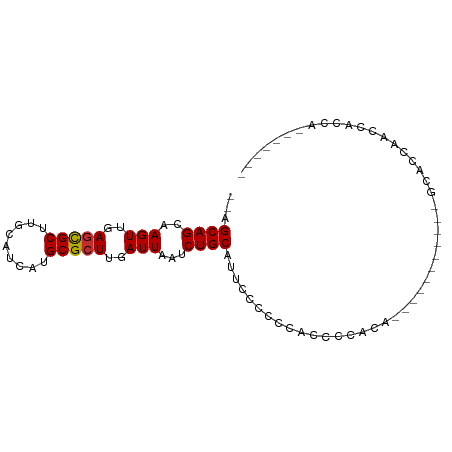
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,453,078 – 11,453,199 |
| Length | 121 |
| Max. P | 0.995440 |

| Location | 11,453,078 – 11,453,182 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.90 |
| Shannon entropy | 0.45240 |
| G+C content | 0.53860 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -18.95 |
| Energy contribution | -19.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.756818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
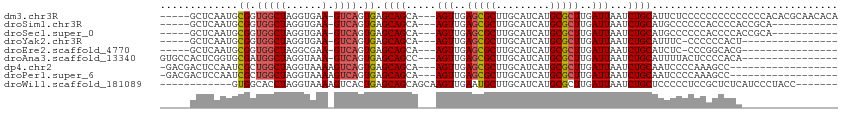
>dm3.chr3R 11453078 104 + 27905053 -----GCUCAAUGCGGUGGCUAGGUGAA-GUCAGUGAGCAGCA---AGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUUCUCCCCCCCCCCCCCCACACGCAACACA -----......((((((((...((.(..-(..((...((((..---(((..(((((.........)))))..)))...))))...))..)..).))...)))).))))..... ( -31.70, z-score = -1.25, R) >droSim1.chr3R 17484425 93 - 27517382 -----GCUCAAUGCGGUGGCUAGGUGAA-GUCAGUGAGCAGCA---AGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUGCCCCCCACCCCACCGCA----------- -----......((((((((...((((..-....(((.((((..---(((..(((((.........)))))..)))...)))).)))....))))))))))))----------- ( -37.00, z-score = -2.38, R) >droSec1.super_0 10622399 93 - 21120651 -----GCUCAAUGCGGUGGCUAGGUGAA-GUCAGUGAGCAGCA---AGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUGCCCCCCACCCCACCGCA----------- -----......((((((((...((((..-....(((.((((..---(((..(((((.........)))))..)))...)))).)))....))))))))))))----------- ( -37.00, z-score = -2.38, R) >droYak2.chr3R 13846424 87 - 28832112 -----GCUCAAUGCGGUGGCUAGGUGAA-GUCAGUGAGCAGCA---AGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUUUC-CCCCCCACU---------------- -----((.....))(((((...((.(((-((......((((..---(((..(((((.........)))))..)))...)))))))))-))..)))))---------------- ( -28.30, z-score = -0.78, R) >droEre2.scaffold_4770 10967917 87 + 17746568 -----GCUCAAUGCGGUGGCUAGGCGAA-GUCAGUGAGCAGCA---AGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUCUC-CCCGGCACG---------------- -----(((.....((.(((((......)-)))).)).((((..---(((..(((((.........)))))..)))...)))).....-...)))...---------------- ( -27.20, z-score = 0.73, R) >droAna3.scaffold_13340 16064019 93 + 23697760 GUGCCACUCGGUGCGAUGGCUAGGUAAA-GUCAGUGAGCAGCC---AGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUUUUACUCCCCACA---------------- (..((....))..)(.(((...((((((-((......((((..---(((..(((((.........)))))..)))...))))))))))))..)))).---------------- ( -29.50, z-score = -0.69, R) >dp4.chr2 30452609 91 + 30794189 -GACGACUCCAAUCGCUGGCUAGGUAAAAGUCAGUGAGCAGCA---AGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAAUCCCCAAAGCC------------------ -...........(((((((((.......)))))))))((((..---(((..(((((.........)))))..)))...)))).............------------------ ( -30.10, z-score = -2.26, R) >droPer1.super_6 5848940 91 + 6141320 -GACGACUCCAAUCGCUGGCUAGGUAAAAGUCAGUGAGCAGCA---AGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAAUCCCCAAAGCC------------------ -...........(((((((((.......)))))))))((((..---(((..(((((.........)))))..)))...)))).............------------------ ( -30.10, z-score = -2.26, R) >droWil1.scaffold_181089 9893865 94 - 12369635 ------------GUGGCACCUAGGUAAAAGUCACUGAGCAGCAGCAAGUUGAAUGCUUGCAUCAUGCGCUUGAUUAAUCUGCUCCCCCUCCGCUCUCAUCCCUACC------- ------------(((((((....))....))))).((((((..((((((.....))))))((((......))))....))))))......................------- ( -23.00, z-score = -0.72, R) >consensus _____GCUCAAUGCGGUGGCUAGGUGAA_GUCAGUGAGCAGCA___AGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUUCCCCCCACCCCA________________ .............((.((((.........)))).)).((((.....(((..(((((.........)))))..)))...))))............................... (-18.95 = -19.20 + 0.25)
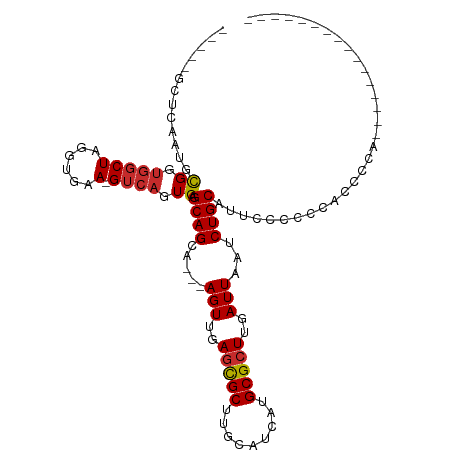
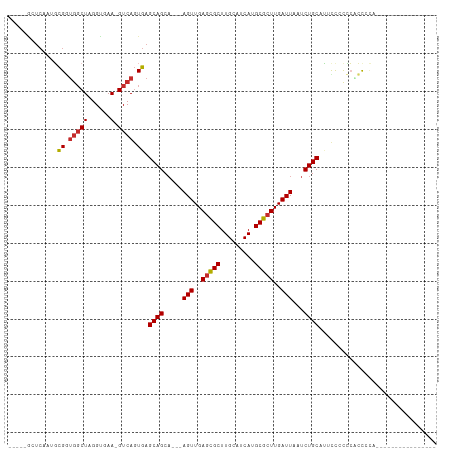
| Location | 11,453,108 – 11,453,199 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 72.87 |
| Shannon entropy | 0.51775 |
| G+C content | 0.55677 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.39 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11453108 91 + 27905053 --AGCAGCAAGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUUCUCCCCCCCCCCCCCCACACGCAACACACCGUGCACUAGCUCCUC --.((((..(((..(((((.........)))))..)))...))))....................((((........))))............ ( -21.00, z-score = -2.71, R) >droSim1.chr3R 17484455 80 - 27517382 --AGCAGCAAGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUGCCCCCCACCCCACC-----------GCACCUAGCACCAGCUCCUC --.((((..(((..(((((.........)))))..)))...)))).................-----------((.....))........... ( -19.60, z-score = -1.53, R) >droSec1.super_0 10622429 80 - 21120651 --AGCAGCAAGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUGCCCCCCACCCCACC-----------GCACCUAGCACCAGCUCCUC --.((((..(((..(((((.........)))))..)))...)))).................-----------((.....))........... ( -19.60, z-score = -1.53, R) >droYak2.chr3R 13846454 77 - 28832112 --AGCAGCAAGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAU-UUCCCCCCCACUCGCA---------GCACCAACCCCCCCUU---- --.((((..(((..(((((.........)))))..)))...))))..-................---------................---- ( -18.30, z-score = -2.74, R) >droEre2.scaffold_4770 10967947 74 + 17746568 --AGCAGCAAGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAU-CUCCCCGGCACGCGCA---------GCACCAGCUCCCC------- --.((((..(((..(((((.........)))))..)))...))))..-.......((....))(---------((....)))....------- ( -20.60, z-score = -0.08, R) >droAna3.scaffold_13340 16064054 71 + 23697760 --AGCAGCCAGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUUUUACUCCCCACAU------------GCCCCCUUCAUA-------- --.((((..(((..(((((.........)))))..)))...))))................------------............-------- ( -18.30, z-score = -2.79, R) >dp4.chr2 30452644 73 + 30794189 --AGCAGCAAGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAAUCCCCAAAGCCCCCA-----------ACCCCAACGCCCA------- --.((((..(((..(((((.........)))))..)))...)))).................-----------.............------- ( -18.30, z-score = -2.86, R) >droPer1.super_6 5848975 73 + 6141320 --AGCAGCAAGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAAUCCCCAAAGCCCCCA-----------ACCCCAACGCCCA------- --.((((..(((..(((((.........)))))..)))...)))).................-----------.............------- ( -18.30, z-score = -2.86, R) >droWil1.scaffold_181089 9893890 86 - 12369635 GCAGCAGCAAGUUGAAUGCUUGCAUCAUGCGCUUGAUUAAUCUGC-UCCCCCUCCGCUCUCAUCCC------UACCCCUCUAACCAUCACUUU ((((..((((((.....))))))((((......))))....))))-....................------..................... ( -14.00, z-score = -1.51, R) >consensus __AGCAGCAAGUUGAGCGCUUGCAUCAUGCGCUUGAUUAAUCUGCAUUCCCCCCACCCCACA___________GCACCAACCACCA_______ ...((((..(((..(((((.........)))))..)))...))))................................................ (-17.38 = -17.39 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:59 2011