| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,440,306 – 11,440,407 |
| Length | 101 |
| Max. P | 0.500000 |

| Location | 11,440,306 – 11,440,407 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.09 |
| Shannon entropy | 0.58281 |
| G+C content | 0.46112 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -10.82 |
| Energy contribution | -10.40 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
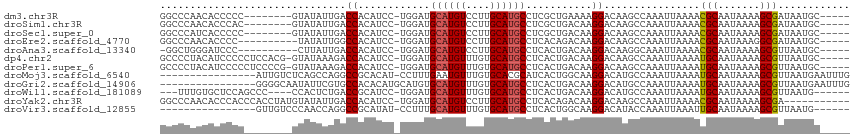
>dm3.chr3R 11440306 101 - 27905053 GGCCCAACACCCCC--------GUAUAUUGACCACAUCC-UGGAUGCAUGUCCUUGCAUGCCUCGCUGAAAAGGACAAGCCAAAUUAAAACGCAAUAAAAGCGAUAAUGC----- (((...........--------((((.....(((.....-))))))).(((((((....((...))....))))))).))).........(((.......))).......----- ( -20.00, z-score = -0.64, R) >droSim1.chr3R 17478245 101 + 27517382 GGCCCAACACCCAC--------GUAUAUUGACCACAUCC-UGGAUGCAUGUCCUUGCAUGCCUCGCUGACAAGGACAAGCCAAAUUAAAACGCAAUAAAAGCGAUAAUGC----- (((...........--------((((.....(((.....-))))))).((((((((((.((...)))).)))))))).))).........(((.......))).......----- ( -23.30, z-score = -1.53, R) >droSec1.super_0 10617080 101 + 21120651 GGCCCAUCACCCCC--------GUAUAUUGACCACAUCC-UGGAUGCAUGUCCUUGCAUGCCUCGCUGACAAGGACAAGCCAAAUUAAAACGCAAUAAAAGCGAUAAUGC----- (((...........--------((((.....(((.....-))))))).((((((((((.((...)))).)))))))).))).........(((.......))).......----- ( -23.30, z-score = -1.44, R) >droEre2.scaffold_4770 10962679 99 - 17746568 GGCCCAACACCCC----------UAUAUUGGCCACAUCC-UGGAUGCAUGUCCUUGCAUGCCUCACAGACAAGGACAAGCCAAAUUAAAACGCAAUAAAGGCGAUAAUGC----- ((((.........----------......))))......-.....((.((((((((..((.....))..)))))))).))..........(((.......))).......----- ( -23.66, z-score = -0.91, R) >droAna3.scaffold_13340 16058985 98 - 23697760 -GGCUGGGAUCCC----------CUUAUUGACCACAUCC-UGGAUGCAUGUCCUUGCAUGCCUCACUGACAAGGACAAGGCAAAUUAAAACGCAAUAAAAGCGUUAAUGC----- -..(..((((...----------............))))-..).(((.((((((((((........)).))))))))..)))......(((((.......))))).....----- ( -25.66, z-score = -1.11, R) >dp4.chr2 30446275 108 - 30794189 GCCCCUACAUCCCCCUCCACG-GUAUAAAGACCACAUCC-UGGAUGCAUGUUUGUGCAUGCCUCACUGACAAGGACAAGCCAAAUUAAAAUGCAAUAAAAGCGUUAAUGC----- ((.............((((.(-(..............))-))))((((((((((.((.(((((........))).)).)))))))....)))))......))........----- ( -19.24, z-score = 0.12, R) >droPer1.super_6 5842600 108 - 6141320 GCCCCUACAUCCCCCUCCCCG-GUAUAAAGACCACAUCC-UGGAUGCAUGUUUGUGCAUGCCUCACUGACAAGGACAAGCCAAAUUAAAAUGCAAUAAAAGCGUUAAUGC----- .......(((((........(-((......)))......-.)))))...(((((.((.(((((........))).)).)))))))...(((((.......))))).....----- ( -18.86, z-score = 0.07, R) >droMoj3.scaffold_6540 22331270 98 - 34148556 ----------------AUUGUCUCAGCCAGGCCGCACAU-CCUUUGAAUGUUUGUGCACGCAUCACUGGCAAGGACAUGCCAAAUUAAAAUGCAAUAAAAGCGUUAAUGAAUUUG ----------------..(((((..(((((((.(((((.-.(.......)..)))))..))....)))))..)))))...((((((..(((((.......)))))....)))))) ( -27.70, z-score = -1.80, R) >droGri2.scaffold_14906 4717829 99 + 14172833 ----------------GGGGCAAUAUUCGUGCCACACAUGCAUGUGCAUGUUUGUGCAUGCCUCACUGACAAGGACAUGCCAAAUUAAAAUGCAAUAAAAGCGUUAAUGAAUUUG ----------------..((((.....(((((((.((((((....)))))).)).)))))(((........)))...))))(((((..(((((.......)))))....))))). ( -27.30, z-score = -0.69, R) >droWil1.scaffold_181089 9888513 101 + 12369635 ---UUUGUGCUCCAGCCC----CCACUCUGACCGCAUCC-UGGAUGCAUGUUUGUGCAUGCCUCACUGACAAGGACAUGCCAAAUUAAAAUGCAAUAAAAGCGUUAAUG------ ---...(((((((((...----................)-)))).))))(((((.((((((((........))).))))))))))...(((((.......)))))....------ ( -25.11, z-score = -1.54, R) >droYak2.chr3R 13840968 103 + 28832112 GGCCCAACACCCACCCACCUAUGUAUAUUGACCACAUCC-UGGAUGCAUGUCCUUGCAUGCCUCACAGACAAGGACAAGCCAAAUUAAAACGCAAUAAAAGCGA----------- (((........((.(((...((((.........))))..-))).))..((((((((..((.....))..)))))))).))).........(((.......))).----------- ( -23.20, z-score = -2.32, R) >droVir3.scaffold_12855 5054251 92 - 10161210 ----------------GUUGUCCCAACCAGGCCGCAUAU-CCUUUGCAUGUUUGUGCAUGCCUCACUGGCAAGGACAUACCAAAUUAAAUUGCAAUAAAAGCGUUAAUG------ ----------------..(((((..........(((((.-.(.......)..))))).((((.....)))).)))))......(((((...((.......)).))))).------ ( -20.10, z-score = -0.14, R) >consensus GGCCCAACACCCCC________GUAUAUUGACCACAUCC_UGGAUGCAUGUCCGUGCAUGCCUCACUGACAAGGACAAGCCAAAUUAAAACGCAAUAAAAGCGUUAAUGC_____ ...............................((..(((....)))((((((....))))))...........))................(((.......)))............ (-10.82 = -10.40 + -0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:58 2011