| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,427,029 – 11,427,090 |
| Length | 61 |
| Max. P | 0.999518 |

| Location | 11,427,029 – 11,427,090 |
|---|---|
| Length | 61 |
| Sequences | 3 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 65.96 |
| Shannon entropy | 0.45208 |
| G+C content | 0.40420 |
| Mean single sequence MFE | -16.37 |
| Consensus MFE | -6.50 |
| Energy contribution | -7.17 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
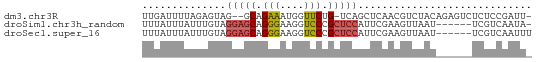
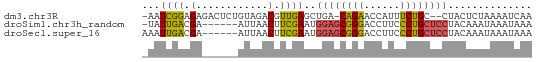
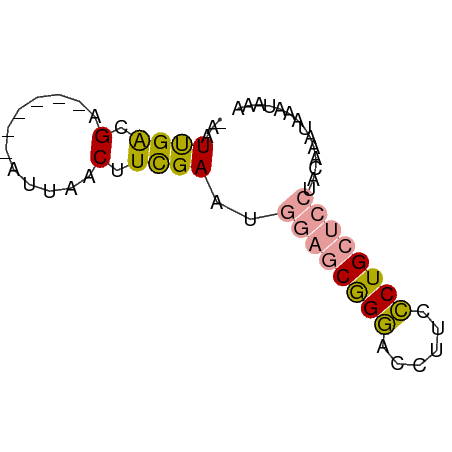
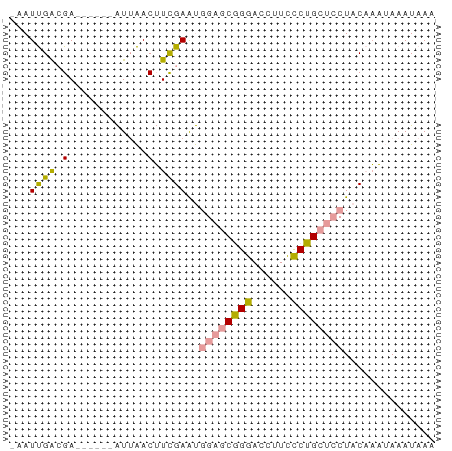
>dm3.chr3R 11427029 61 + 27905053 UUGAUUUUAGAGUAG--GCAGAAAUGGUUCUG-UCAGCUCAACGUCUACAGAGUCUCUCCGAUU- ..((((((.((((.(--((((((....)))))-)).))))...(....))))))).........- ( -15.70, z-score = -1.22, R) >droSim1.chr3h_random 100549 58 - 1452968 UUUAUUUAUUUGUAGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAU------UCGUCAAUA- ..............(((((.(((.....))))))))...((((.....)------)))......- ( -16.70, z-score = -3.15, R) >droSec1.super_16 1851330 59 - 1878335 UUUAUUUAUUUGUAGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAU------UCGUCAAUUU ..............(((((.(((.....))))))))...((((.....)------)))....... ( -16.70, z-score = -3.18, R) >consensus UUUAUUUAUUUGUAGGAGCAGGGAAGGUCCCGCUCCAUUCGAAGUUAAU______UCGUCAAUU_ ..............(((((.(((....))).)))))............................. ( -6.50 = -7.17 + 0.67)
| Location | 11,427,029 – 11,427,090 |
|---|---|
| Length | 61 |
| Sequences | 3 |
| Columns | 65 |
| Reading direction | reverse |
| Mean pairwise identity | 65.96 |
| Shannon entropy | 0.45208 |
| G+C content | 0.40420 |
| Mean single sequence MFE | -15.63 |
| Consensus MFE | -9.86 |
| Energy contribution | -10.53 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
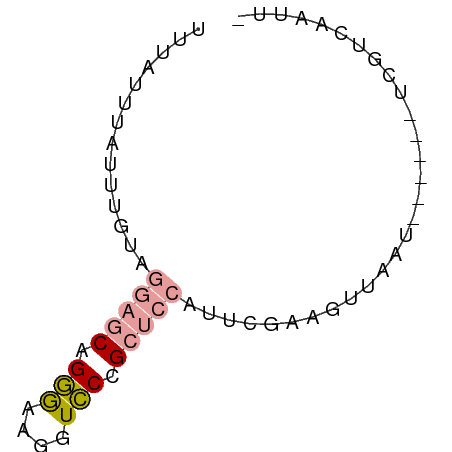
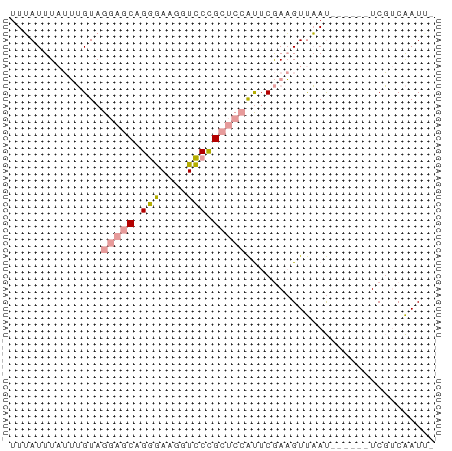
>dm3.chr3R 11427029 61 - 27905053 -AAUCGGAGAGACUCUGUAGACGUUGAGCUGA-CAGAACCAUUUCUGC--CUACUCUAAAAUCAA -...(((((...)))))..((..(((((..(.-(((((....))))).--)..))).))..)).. ( -11.10, z-score = -0.24, R) >droSim1.chr3h_random 100549 58 + 1452968 -UAUUGACGA------AUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCUACAAAUAAAUAAA -..(((.(((------(.....))))...((((((((......))))))))..)))......... ( -17.90, z-score = -5.16, R) >droSec1.super_16 1851330 59 + 1878335 AAAUUGACGA------AUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCUACAAAUAAAUAAA ...(((.(((------(.....))))...((((((((......))))))))..)))......... ( -17.90, z-score = -5.12, R) >consensus _AAUUGACGA______AUUAACUUCGAAUGGAGCGGGACCUUCCCUGCUCCUACAAAUAAAUAAA .............................((((((((......)))))))).............. ( -9.86 = -10.53 + 0.67)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:55 2011