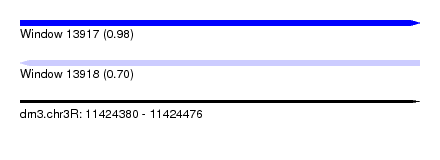
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,424,380 – 11,424,476 |
| Length | 96 |
| Max. P | 0.982376 |

| Location | 11,424,380 – 11,424,476 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.44 |
| Shannon entropy | 0.50291 |
| G+C content | 0.39679 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.17 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
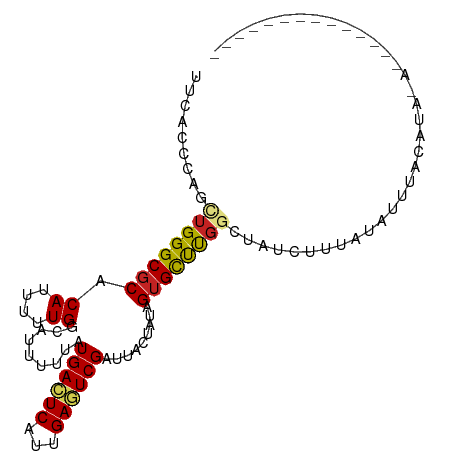
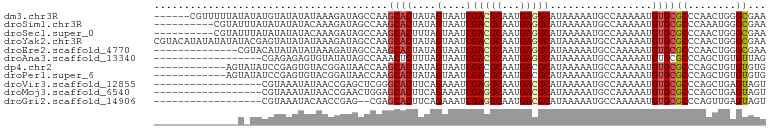
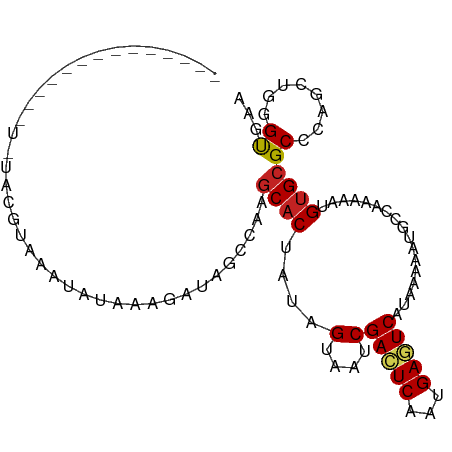
>dm3.chr3R 11424380 96 + 27905053 UUCGCCCAGUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUAUACAUAUAUAAAAACG------ .......(((..(((((.((....)).........((((((...)))))).........)))))..)))...((((((((....))))))))....------ ( -22.50, z-score = -2.40, R) >droSim1.chr3R 17469919 92 - 27517382 UUCGCCCAUUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUGUAUAUAUAUAAAUACG---------- ..(((((....))))).(((....((((.......((((((...))))))..(((....)))....))))....)))...............---------- ( -20.10, z-score = -1.70, R) >droSec1.super_0 10608715 92 - 21120651 UUCGCCCAGUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAAAGUGCUUGGCUAUCUUUGUAUAUAUAUAAAUACG---------- ...((((....))))((.((.....((((((((..((((((...))))))......)))))))))))).......((((........)))).---------- ( -22.10, z-score = -1.97, R) >droYak2.chr3R 13832009 102 - 28832112 UUCGCCCAGUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUAUACUCGUAUAUAUAUAUGUACG .......(((..(((((.((....)).........((((((...)))))).........)))))..))).....((((((((....))))))))........ ( -24.20, z-score = -1.88, R) >droEre2.scaffold_4770 10954551 88 + 17746568 UUCGCCCAGUUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUAUAUGUACG-------------- ..(((((....)))))..........((((...(((((.......((((((.(((....))).))))))....)))))...))))...-------------- ( -20.10, z-score = -1.40, R) >droAna3.scaffold_13340 16050241 84 + 23697760 CUAACACAGCUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAAAGAGUUUGGCUAUAUACACUCUCUCG------------------ .......(((..(((((.((....)))).......((((((...))))))...........)))..)))...............------------------ ( -17.70, z-score = -1.86, R) >dp4.chr2 30434386 90 + 30794189 CACACACAGCUGGGCGCACAUUUUUGGCAUUUUUAUGAGUCAUUGAGUCGAUUACUAUAGUGCUUGGUUAUCCGUACACUCGGAUAUACU------------ .......(((..(((((..........(((....)))(((.((((...)))).)))...)))))..)))(((((......))))).....------------ ( -20.50, z-score = -1.15, R) >droPer1.super_6 5830214 90 + 6141320 CACACACAGCUGGGCGCACAUUUUUGGCAUUUUUAUGAGUCAUUGAGUCGAUUACUAUAGUGCUUGGUUAUCCGUACACUCGGAUAUACU------------ .......(((..(((((..........(((....)))(((.((((...)))).)))...)))))..)))(((((......))))).....------------ ( -20.50, z-score = -1.15, R) >droVir3.scaffold_12855 5043534 84 + 10161210 ACUACUCAGCUGGGCGCACAUUUUUGGCAUUUUUAUGAGUCAUUGACUCGAUUUCUGAAGUGCCCGAGCUCGGUUAUAUUUACG------------------ ...(((.((((((((((.((....)).........((((((...)))))).........)))))).)))).)))..........------------------ ( -24.30, z-score = -2.73, R) >droMoj3.scaffold_6540 22321365 84 + 34148556 ACUACUCAGCUGGGCGCACAUUUUUGGCAUUUUUAUGAGUCAUUGACUCGAUUUCUGAAGUGCUCCAGUUCGGUUAUAUUUACG------------------ ...(((.((((((((((.((....)).........((((((...)))))).........)))).)))))).)))..........------------------ ( -21.90, z-score = -2.44, R) >droGri2.scaffold_14906 4707424 82 - 14172833 ACUACUCAACUGGGCGCACAUUUUUGGCAUUUUUAUGAGUCAUUGACUCGAUUUCUGAAGUGCUCG--CUCGGUUGUAUUUACG------------------ ......((((((((((.........((((((((..((((((...))))))......))))))))))--))))))))........------------------ ( -25.10, z-score = -3.90, R) >consensus UUCACCCAGCUGGGCGCACAUUUUUGGCAUUUUUAUGACUCAUUGAGUCGAUUACUAUAGUGCUUGGCUAUCUUUAUAUUUACAUA_A______________ .........((((((((.((....)).........((((((...)))))).........))))))))................................... (-12.32 = -12.17 + -0.15)

| Location | 11,424,380 – 11,424,476 |
|---|---|
| Length | 96 |
| Sequences | 11 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 74.44 |
| Shannon entropy | 0.50291 |
| G+C content | 0.39679 |
| Mean single sequence MFE | -19.08 |
| Consensus MFE | -8.27 |
| Energy contribution | -8.09 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.700513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11424380 96 - 27905053 ------CGUUUUUAUAUAUGUAUAUAUAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAACUGGGCGAA ------.(((((((((((....)))))))))))......((((....(....)(((((...))))).................))))((((....))))... ( -24.00, z-score = -3.06, R) >droSim1.chr3R 17469919 92 + 27517382 ----------CGUAUUUAUAUAUAUACAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAAAUGGGCGAA ----------.((((((.(((...(((....((((.......)))).)))...(((((...)))))))).))))))..........(((((....))))).. ( -20.30, z-score = -2.43, R) >droSec1.super_0 10608715 92 + 21120651 ----------CGUAUUUAUAUAUAUACAAAGAUAGCCAAGCACUUUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAACUGGGCGAA ----------.((((((.(((...(((((((...((...)).)))).)))...(((((...)))))))).))))))..........(((((....))))).. ( -20.70, z-score = -2.36, R) >droYak2.chr3R 13832009 102 + 28832112 CGUACAUAUAUAUAUACGAGUAUAUAUAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAACUGGGCGAA .((((((.((((((((....))))))))...........((((....))....(((((...)))))........)).....))))))((((....))))... ( -25.70, z-score = -3.34, R) >droEre2.scaffold_4770 10954551 88 - 17746568 --------------CGUACAUAUAUAUAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAACUGGGCGAA --------------.((((((.........(((.((...........)).)))(((((...)))))...............))))))((((....))))... ( -21.00, z-score = -2.83, R) >droAna3.scaffold_13340 16050241 84 - 23697760 ------------------CGAGAGAGUGUAUAUAGCCAAACUCUUUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAGCUGUGUUAG ------------------..(((((((............)))))))(((....(((((...)))))........((((....)).)).....)))....... ( -13.70, z-score = -0.49, R) >dp4.chr2 30434386 90 - 30794189 ------------AGUAUAUCCGAGUGUACGGAUAACCAAGCACUAUAGUAAUCGACUCAAUGACUCAUAAAAAUGCCAAAAAUGUGCGCCCAGCUGUGUGUG ------------....((((((......)))))).....((((.(((((....((.((...)).))........((((....)).)).....))))))))). ( -16.10, z-score = -0.19, R) >droPer1.super_6 5830214 90 - 6141320 ------------AGUAUAUCCGAGUGUACGGAUAACCAAGCACUAUAGUAAUCGACUCAAUGACUCAUAAAAAUGCCAAAAAUGUGCGCCCAGCUGUGUGUG ------------....((((((......)))))).....((((.(((((....((.((...)).))........((((....)).)).....))))))))). ( -16.10, z-score = -0.19, R) >droVir3.scaffold_12855 5043534 84 - 10161210 ------------------CGUAAAUAUAACCGAGCUCGGGCACUUCAGAAAUCGAGUCAAUGACUCAUAAAAAUGCCAAAAAUGUGCGCCCAGCUGAGUAGU ------------------..........((..((((.((((............(((((...)))))........((((....)).))))))))))..))... ( -21.50, z-score = -2.46, R) >droMoj3.scaffold_6540 22321365 84 - 34148556 ------------------CGUAAAUAUAACCGAACUGGAGCACUUCAGAAAUCGAGUCAAUGACUCAUAAAAAUGCCAAAAAUGUGCGCCCAGCUGAGUAGU ------------------................((((.((((....(....)(((((...))))).................))))..))))......... ( -15.90, z-score = -1.24, R) >droGri2.scaffold_14906 4707424 82 + 14172833 ------------------CGUAAAUACAACCGAG--CGAGCACUUCAGAAAUCGAGUCAAUGACUCAUAAAAAUGCCAAAAAUGUGCGCCCAGUUGAGUAGU ------------------........((((.(.(--((.(((.....(....)(((((...)))))................))).))).).))))...... ( -14.90, z-score = -1.08, R) >consensus ______________U_UACGUAAAUAUAAAGAUAGCCAAGCACUAUAGUAAUCGACUCAAUGAGUCAUAAAAAUGCCAAAAAUGUGCGCCCAGCUGGGUGAA .......................................((((....(....)(((((...))))).................))))((........))... ( -8.27 = -8.09 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:54 2011