| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,396,812 – 11,396,941 |
| Length | 129 |
| Max. P | 0.500000 |

| Location | 11,396,812 – 11,396,941 |
|---|---|
| Length | 129 |
| Sequences | 4 |
| Columns | 129 |
| Reading direction | forward |
| Mean pairwise identity | 69.22 |
| Shannon entropy | 0.50247 |
| G+C content | 0.44965 |
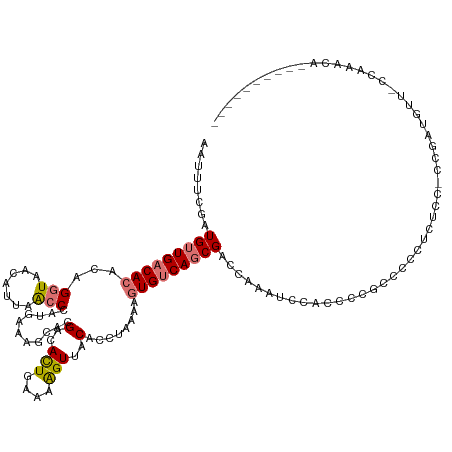
| Mean single sequence MFE | -19.50 |
| Consensus MFE | -9.09 |
| Energy contribution | -11.15 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
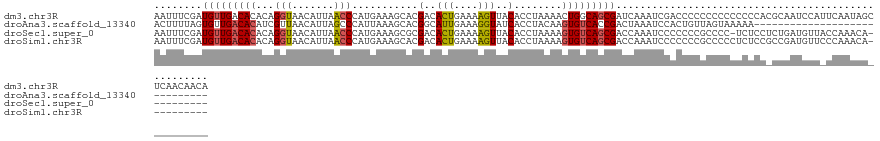
>dm3.chr3R 11396812 129 + 27905053 AAUUUCGAUGUUGACACACAGGUAACAUUAACCCAUGAAAGCACGACACUGAAAAGUUACACCUAAAACUGGCAGCGAUCAAAUCGACCCCCCCCCCCCCCACGCAAUCCAUUCAAUAGCUCAACAACA ......(.(((((((.((..(((.......)))..))...((......(((...((((........))))..)))((((...)))).................)).............).)))))).). ( -16.50, z-score = -0.56, R) >droAna3.scaffold_13340 16019961 100 + 23697760 ACUUUUAGUGUUGACACAUCGUUAACAUUAGCCCAUUAAAGCACGGCAUUGAAAGGUAUCACCUACAAGUGUCACCGACUAAAUCCACUGUUAGUAAAAA----------------------------- .....((((((((((.....))))))))))((........))..((((((...(((.....)))...))))))....(((((........))))).....----------------------------- ( -21.40, z-score = -1.81, R) >droSec1.super_0 10581326 118 - 21120651 AAUUUCGAUGUUGACACACAGGUAACAUUAACCCAUGAAAGCGCGACACUGAAAAGUUACACCUAAAAGUGUCAGCGACCAAAUCCCCCCCGCCCC-UCUCCUCUGAUGUUACCAAACA---------- ....................((((((((((..............((((((....((......))...)))))).(((.............)))...-.......)))))))))).....---------- ( -22.32, z-score = -2.22, R) >droSim1.chr3R 17439378 119 - 27517382 AAUUUCGAUGUUGACACACAGGUAACAUUAACCCAUGAAAGCACGACACUGAAAAGUUACACCUAAAAGUGUCAGCGACCAAAUCCCCCCCGCCCCCUCUCCGCCGAUGUUCCCAAACA---------- ....(((.(((((((((..((((..(((......)))..........(((....)))...))))....)))))))))..............((.........)))))............---------- ( -17.80, z-score = -0.98, R) >consensus AAUUUCGAUGUUGACACACAGGUAACAUUAACCCAUGAAAGCACGACACUGAAAAGUUACACCUAAAAGUGUCAGCGACCAAAUCCACCCCGCCCCCUCUCC_CCGAUGUU_CCAAACA__________ ........(((((((((...(((.......)))...........(..(((....)))..)........))))))))).................................................... ( -9.09 = -11.15 + 2.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:49 2011