| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,383,185 – 11,383,291 |
| Length | 106 |
| Max. P | 0.986576 |

| Location | 11,383,185 – 11,383,291 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.35 |
| Shannon entropy | 0.60591 |
| G+C content | 0.47573 |
| Mean single sequence MFE | -32.29 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.41 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
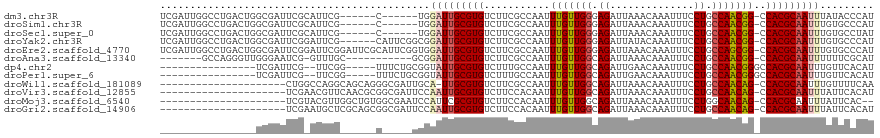
>dm3.chr3R 11383185 106 + 27905053 UCGAUUGGCCUGACUGGCGAUUCGCAUUCG------C------UGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGG-CCACGCAAUUUAUACCCAU ..(..(((((....((((((..((((.((.------.------..)).)))).....))))))..(((..((((((((.....))))))))..))).))-)))..)............. ( -35.10, z-score = -2.59, R) >droSim1.chr3R 17425802 106 - 27517382 UCGAUUGGCCUGACUGGCGAUUCGCAUUCG------C------UGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGG-CCACGCAAUUUGUGCCCAU .....(((((....((((((..((((.((.------.------..)).)))).....))))))..(((..((((((((.....))))))))..))).))-))).(((.....))).... ( -36.00, z-score = -2.02, R) >droSec1.super_0 10567703 106 - 21120651 UCGAUUGGCCUGACUGGCGAUUCGCAUUCG------C------UGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAACGG-CCACGCAAUUUGUGCCUAU .....(((((....((((((..((((.((.------.------..)).)))).....))))))..(((..((((((((.....))))))))..))).))-))).(((.....))).... ( -36.00, z-score = -2.28, R) >droYak2.chr3R 13788869 112 - 28832112 UCGAUUGGCCUGACUGGCGAUUCGGAUUCG------CAUUCGGCGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAUACAAAUUUCCUGCCAACGG-CCACGCAAUUUGUGCCCAU .....(((((....((((((...((((.((------((.((....)).)))).))))))))))..(((..((((((((.....))))))))..))).))-))).(((.....))).... ( -39.60, z-score = -1.83, R) >droEre2.scaffold_4770 10911323 118 + 17746568 UCGAUUGGCCUGACUGGCGAUUCGGAUUCGGAUUCGCAUUCGGUGGAUUGCGUGUCUUCGCCAAUUUGUUGGGAGAUUAAACAAAUUUCCUGCCAGCGG-CCACGCAAUUUGUGCCCAU .....(((((.(.(((((.....((....((((.((((.((....)).)))).))))...))........((((((((.....))))))))))))))))-))).(((.....))).... ( -39.70, z-score = -0.93, R) >droAna3.scaffold_13340 16006167 99 + 23697760 -------GCCAGGGUUGGGAAUCG-GUUUGC-----------GCGGAUUGCGUGUCUUCGCCAAUUUGUUGGCAGAUUAAACAAAUUUCCUGCCAACGG-CCACGCAAUUUUUUCGCAU -------.((......))......-...(((-----------(.((((((((((.....(((.....((((((((..............))))))))))-)))))))))))...)))). ( -31.44, z-score = -0.81, R) >dp4.chr2 30386439 96 + 30794189 ----------------UCGAUUCG--UUCGG-----UUUCUGCGGUAUUGCGUGUCUUUGCCAAUUUGUUGGCAGAUUGAACAAAUUUCCUGCCAACGGGCCACGCAAUUUGUUCACAU ----------------.(((....--.)))(-----(....((((.((((((((.....(((....(((((((((...(((.....)))))))))))))))))))))))))))..)).. ( -28.80, z-score = -1.65, R) >droPer1.super_6 5780424 96 + 6141320 ----------------UCGAUUCG--UUCGG-----UUUCUGCGGUAUUGCGUGUCUUUGCCAAUUUGUUGGCAGAUUGAACAAAUUUCCUGCCAACGGGCCACGCAAUUUGUUCACAU ----------------.(((....--.)))(-----(....((((.((((((((.....(((....(((((((((...(((.....)))))))))))))))))))))))))))..)).. ( -28.80, z-score = -1.65, R) >droWil1.scaffold_181089 9836315 96 - 12369635 ---------------------CUGGCCAGGCAGCAGGGCGAUUGCA-UUGCGUGUCUUCGCCAAUUUGUUGGCAGAUUAAACAAAUUUCCUGCCAACAG-CCACGCAAUUUGUUUUCAA ---------------------...((..(((.....(((((..(((-(...))))..)))))....(((((((((..............))))))))))-))..))............. ( -31.94, z-score = -1.75, R) >droVir3.scaffold_12855 4984106 97 + 10161210 ---------------------UCGAACGUUCAACGCGGCGAUUCCAAUUGCGUGUCUUCCACAAUUUGUUGGCAGAUUAAACAAAUUUCCUGCCAACAG-CCACGCAAUUUAUUCACAU ---------------------..(((((((......)))).))).(((((((((.((..........((((((((..............))))))))))-.)))))))))......... ( -24.94, z-score = -2.76, R) >droMoj3.scaffold_6540 22272187 95 + 34148556 ---------------------UCGUACGUUGGCUGUGGCGAAUCCAUUCGCGUGUCUUCCACAAUUUGUUGGCAGAUUAAACAAAUUUCCUGGCAACAG-CCACGCAAUUUAUUCAC-- ---------------------.....(((.((((((.(((((....)))))(((.....)))....(((..(.(((((.....))))).)..)))))))-)))))............-- ( -25.90, z-score = -1.87, R) >droGri2.scaffold_14906 4654870 97 - 14172833 ---------------------UCGAAUGCUCGCAGCGGCGAUUCCAAUUGCGUGUCUUCCACAAUUUGUUGGCAGAUUAAACAAAUUUCCUGCCAACAG-CCACGCAAUUUAUUCACAU ---------------------..(((((.((((....))))....(((((((((.((..........((((((((..............))))))))))-.)))))))))))))).... ( -29.24, z-score = -3.59, R) >consensus ________________GCGAUUCG_AUUCG______C_____UCGGAUUGCGUGUCUUCGCCAAUUUGUUGGCAGAUUAAACAAAUUUCCUGCCAACGG_CCACGCAAUUUGUUCACAU .............................................(((((((((...........(((((((.((..............)).)))))))..)))))))))......... (-15.37 = -15.41 + 0.04)
| Location | 11,383,185 – 11,383,291 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.35 |
| Shannon entropy | 0.60591 |
| G+C content | 0.47573 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -14.17 |
| Energy contribution | -14.12 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
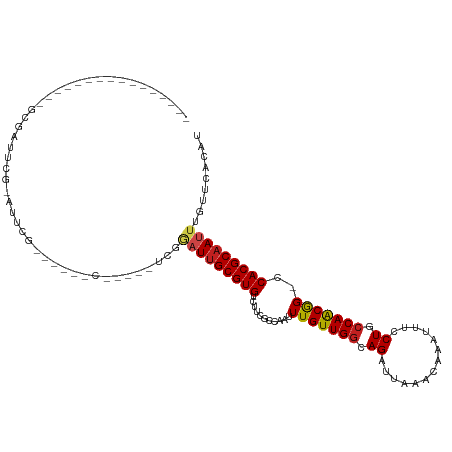
>dm3.chr3R 11383185 106 - 27905053 AUGGGUAUAAAUUGCGUGG-CCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCA------G------CGAAUGCGAAUCGCCAGUCAGGCCAAUCGA ....(((.....))).(((-(((((((.(((............))).)))))..((((((((.....((((.((..------.------.)).))))..))))))))..)))))..... ( -31.70, z-score = -0.95, R) >droSim1.chr3R 17425802 106 + 27517382 AUGGGCACAAAUUGCGUGG-CCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCA------G------CGAAUGCGAAUCGCCAGUCAGGCCAAUCGA ..((.(((.......))).-))((((((..((((((((((.........))))))))(((((.....((((.((..------.------.)).))))..)))))..))..))))))... ( -32.90, z-score = -1.01, R) >droSec1.super_0 10567703 106 + 21120651 AUAGGCACAAAUUGCGUGG-CCGUUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCA------G------CGAAUGCGAAUCGCCAGUCAGGCCAAUCGA ....(((.....))).(((-(((((((.(((............))).)))))..((((((((.....((((.((..------.------.)).))))..))))))))..)))))..... ( -32.80, z-score = -1.51, R) >droYak2.chr3R 13788869 112 + 28832112 AUGGGCACAAAUUGCGUGG-CCGUUGGCAGGAAAUUUGUAUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCGCCGAAUG------CGAAUCCGAAUCGCCAGUCAGGCCAAUCGA ....(((.....))).(((-(((((((.(((..((....))..))).)))))..((((((((.((..((((.((....)).))------))..))....))))))))..)))))..... ( -34.10, z-score = -0.76, R) >droEre2.scaffold_4770 10911323 118 - 17746568 AUGGGCACAAAUUGCGUGG-CCGCUGGCAGGAAAUUUGUUUAAUCUCCCAACAAAUUGGCGAAGACACGCAAUCCACCGAAUGCGAAUCCGAAUCCGAAUCGCCAGUCAGGCCAAUCGA ....(((.....))).(((-((((((((.(((....(((((....((((((....)))).)))))))((((.((....)).))))..)))((.......))))))))..)))))..... ( -34.30, z-score = -0.59, R) >droAna3.scaffold_13340 16006167 99 - 23697760 AUGCGAAAAAAUUGCGUGG-CCGUUGGCAGGAAAUUUGUUUAAUCUGCCAACAAAUUGGCGAAGACACGCAAUCCGC-----------GCAAAC-CGAUUCCCAACCCUGGC------- .((((.....(((((((((-(((((((((((............))))))))).....))).....))))))))...)-----------)))...-......(((....))).------- ( -30.20, z-score = -1.39, R) >dp4.chr2 30386439 96 - 30794189 AUGUGAACAAAUUGCGUGGCCCGUUGGCAGGAAAUUUGUUCAAUCUGCCAACAAAUUGGCAAAGACACGCAAUACCGCAGAAA-----CCGAA--CGAAUCGA---------------- .((((.....(((((((((((.(((((((((((.....)))...)))))))).....))).....))))))))..))))....-----.(((.--....))).---------------- ( -26.90, z-score = -1.58, R) >droPer1.super_6 5780424 96 - 6141320 AUGUGAACAAAUUGCGUGGCCCGUUGGCAGGAAAUUUGUUCAAUCUGCCAACAAAUUGGCAAAGACACGCAAUACCGCAGAAA-----CCGAA--CGAAUCGA---------------- .((((.....(((((((((((.(((((((((((.....)))...)))))))).....))).....))))))))..))))....-----.(((.--....))).---------------- ( -26.90, z-score = -1.58, R) >droWil1.scaffold_181089 9836315 96 + 12369635 UUGAAAACAAAUUGCGUGG-CUGUUGGCAGGAAAUUUGUUUAAUCUGCCAACAAAUUGGCGAAGACACGCAA-UGCAAUCGCCCUGCUGCCUGGCCAG--------------------- ................(((-(((((((((((............))))))))).....((((.......(((.-.((....))..))))))).))))).--------------------- ( -26.71, z-score = -0.28, R) >droVir3.scaffold_12855 4984106 97 - 10161210 AUGUGAAUAAAUUGCGUGG-CUGUUGGCAGGAAAUUUGUUUAAUCUGCCAACAAAUUGUGGAAGACACGCAAUUGGAAUCGCCGCGUUGAACGUUCGA--------------------- ...(((((.....((((((-(((((((((((............))))))))))(((((((.......)))))))......))))))).....))))).--------------------- ( -34.90, z-score = -3.43, R) >droMoj3.scaffold_6540 22272187 95 - 34148556 --GUGAAUAAAUUGCGUGG-CUGUUGCCAGGAAAUUUGUUUAAUCUGCCAACAAAUUGUGGAAGACACGCGAAUGGAUUCGCCACAGCCAACGUACGA--------------------- --..........(((((((-((((.((.((...((((((....(((.(((........))).)))...))))))...)).)).)))))).)))))...--------------------- ( -26.90, z-score = -1.28, R) >droGri2.scaffold_14906 4654870 97 + 14172833 AUGUGAAUAAAUUGCGUGG-CUGUUGGCAGGAAAUUUGUUUAAUCUGCCAACAAAUUGUGGAAGACACGCAAUUGGAAUCGCCGCUGCGAGCAUUCGA--------------------- ...(((((...((((((((-(((((((((((............))))))))))(((((((.......)))))))......))))).))))..))))).--------------------- ( -35.00, z-score = -3.19, R) >consensus AUGGGAACAAAUUGCGUGG_CCGUUGGCAGGAAAUUUGUUUAAUCUGCCAACAAAUUGGCGAAGACACGCAAUCCGA_____G______CGAAU_CGAAUCGC________________ ..........((((((((..((.......)).((((((((.........))))))))........)))))))).............................................. (-14.17 = -14.12 + -0.05)

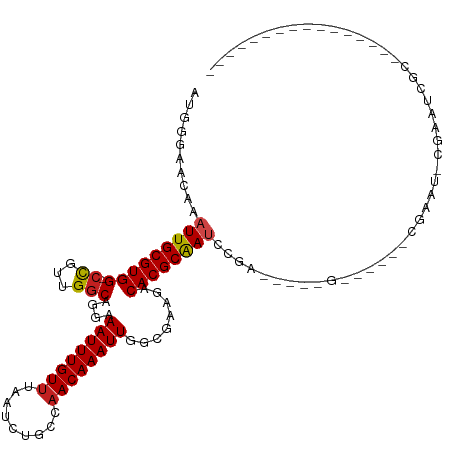
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:47 2011