| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,381,904 – 11,382,050 |
| Length | 146 |
| Max. P | 0.703592 |

| Location | 11,381,904 – 11,382,050 |
|---|---|
| Length | 146 |
| Sequences | 6 |
| Columns | 162 |
| Reading direction | forward |
| Mean pairwise identity | 63.93 |
| Shannon entropy | 0.65599 |
| G+C content | 0.35652 |
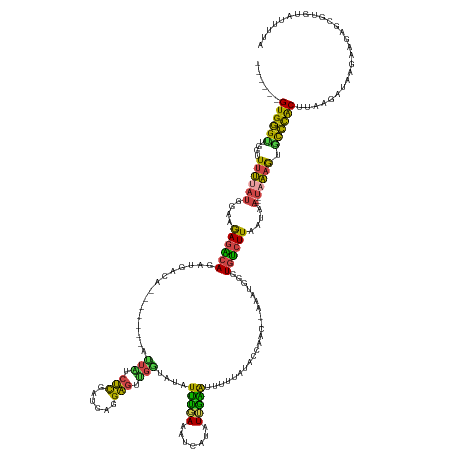
| Mean single sequence MFE | -34.37 |
| Consensus MFE | -13.95 |
| Energy contribution | -11.41 |
| Covariance contribution | -2.55 |
| Combinations/Pair | 1.91 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.703592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
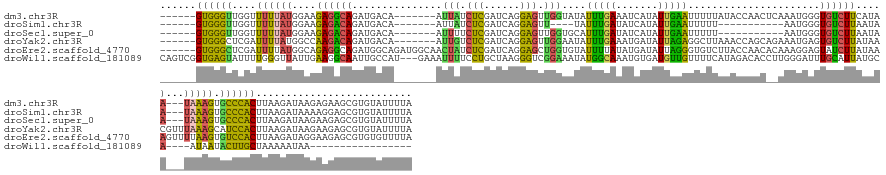
>dm3.chr3R 11381904 146 + 27905053 ------GUGGGUUGGUUUUUAUGGAAGAGGCAGAUGACA-------AUUAUCUCGAUCAGGAGUUGGUAUAUUUGAAAUCAUAUUGAAUUUUUAUACCAACUCAAAUGGGUGUCUUCAUAA---UAAAGUGCCCACUUAAGAUAAGAGAAGCGUGUAUUUUA ------((((((....((((((.(((((.((((((((..-------.))))))...(((.(((((((((((...(((.((.....)).))).)))))))))))...))))).)))))...)---))))).)))))).(((((((.(.....)...))))))) ( -43.30, z-score = -3.86, R) >droSim1.chr3R 17424492 131 - 27517382 ------GUGGGUUGGUUUUUAUGGAAGAGACAGAUGACA-------AUUAUCUCGAUCAGGAGUU----UAUUUGAUAUCAUAUUGAAUUUUU-----------AAUGGGUGUCUUAAUAA---UAAAGUGCCCACUUAAGAUAAAAGGAGCGUGUAUUUUA ------((((((....((((((....((((((((((.((-------(.((.(((......)))..----)).))).)))).((((((....))-----------))))..))))))....)---))))).)))))).......................... ( -27.20, z-score = -0.79, R) >droSec1.super_0 10566473 135 - 21120651 ------GUGGGUUGGUUUUUAUGGAAGAGACAGAUGACA-------AUUUUCUCGAUCAGGAGUUGGUGCAUUUGAUAUCAUAUUGAAUUUUU-----------AAUGGGUGUCUUAAUAA---UAAAGUGCCCACUUAAGAUAAGAAGAGCGUGUAUUUUA ------....(((.((((((.....)))))).))).(((-------.((((((..(((..((((.((.((((((((((((.((((((....))-----------)))))))))).......---..))))))))))))..))).))))))...)))...... ( -28.60, z-score = -0.53, R) >droYak2.chr3R 13787563 149 - 28832112 ------GUGGGCUCGAUUUUAUGGCCAAGACAGAUGACA-------AUUGUCUCGAUCAGGAGUUGGAAUAUUUGAAAUGAUAUUAGAGGCUUAAACCAGCAGAAAUGAGUGUCUUAUAACGUUUAAAGCAUCCACUUAAGAUAAGAAGAGCGUGUAUUUUA ------....((((..(((((((..(.((((((......-------.)))))).)..)..((((.(((...((((((.((.(((.(((.(((((............))))).))).))).))))))))...)))))))...)))))).)))).......... ( -31.30, z-score = 0.03, R) >droEre2.scaffold_4770 10910088 156 + 17746568 ------GUGGGCUCGAUUUUAUGGCAGAGGCAGAUGGCAGAUGGCAACUAUCUCGAUCAGGAGCUGGUGUAUUUUAUAUGAUAUUAGGGUGUCUUACCAACACAAAGGAGUAUCUUAUAAAGUUUUAAGUGUCCACUUAAGAUAGGAAGAGCGUGUGUUUUA ------....((((.......((.((...((.....))...)).)).((((((..(...((((((.....((((((((.((((((...((((.......)))).....)))))).))))))))....))).)))..)..))))))...)))).......... ( -37.60, z-score = 0.08, R) >droWil1.scaffold_181089 9834943 138 - 12369635 CAGUCGGUGAGUAUUUUGGGUUAUUGAAGGCAAUUGCCAU---GAAAUUUUCCUGCUAAGGGUCGGAAAUAUGGCAAAUGUGAUGUUGUUUUCAUAGACACCUUGGGAUUUGCAUUAUGCA----AUAAUACUUGCUAAAAAUAA----------------- .....((..((((((..(((((..((((((((((((((((---(....(((((.((.....)).))))))))))))........))))))))))..))).)).......((((.....)))----).))))))..))........----------------- ( -38.20, z-score = -2.16, R) >consensus ______GUGGGUUGGUUUUUAUGGAAGAGACAGAUGACA_______AUUAUCUCGAUCAGGAGUUGGUAUAUUUGAAAUCAUAUUGAAUUUUUAUACCAAC__AAAUGGGUGUCUUAAUAA___UAAAGUGCCCACUUAAGAUAAGAAGAGCGUGUAUUUUA ......((((((..............((((((...............(((.(((......))).)))....(((((.......)))))......................))))))..............)))))).......................... (-13.95 = -11.41 + -2.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:46 2011