
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,331,344 – 11,331,441 |
| Length | 97 |
| Max. P | 0.789202 |

| Location | 11,331,344 – 11,331,441 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 67.19 |
| Shannon entropy | 0.64030 |
| G+C content | 0.46617 |
| Mean single sequence MFE | -19.44 |
| Consensus MFE | -7.57 |
| Energy contribution | -8.32 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.789202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
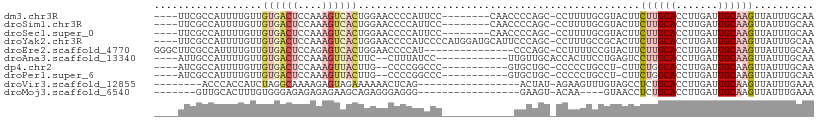
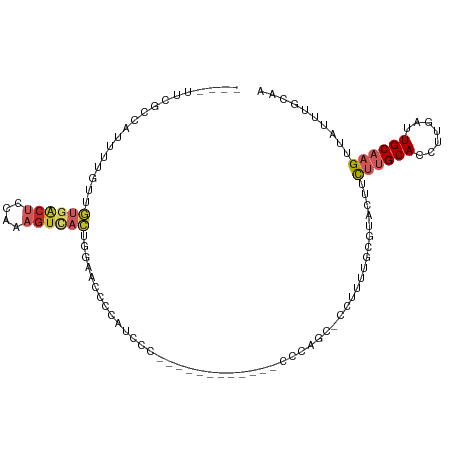
>dm3.chr3R 11331344 97 - 27905053 ----UUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUCC--------CAACCCCAGC-CCUUUUGCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAA ----...((.........((((((....)))))).((((.....))))--------........))-....(((((.....((((((.......)))))).....))))) ( -20.20, z-score = -2.52, R) >droSim1.chr3R 17372511 97 + 27517382 ----UUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUCC--------CAACCCCAGC-CCUUUUGCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAA ----...((.........((((((....)))))).((((.....))))--------........))-....(((((.....((((((.......)))))).....))))) ( -20.20, z-score = -2.52, R) >droSec1.super_0 10507317 97 + 21120651 ----UUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUCC--------CAACCCCAGC-CCUUUUGCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAA ----...((.........((((((....)))))).((((.....))))--------........))-....(((((.....((((((.......)))))).....))))) ( -20.20, z-score = -2.52, R) >droYak2.chr3R 13735616 105 + 28832112 ----UUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUCCCCAUGGAUGCAUUCCCAGC-CCUUUGCCGCACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAA ----...((.........((((((....)))))).((((...(((((....)))))..))))..))-........(((...((((((.......)))))).....))).. ( -25.30, z-score = -2.27, R) >droEre2.scaffold_4770 10859019 94 - 17746568 GGGCUUCGCCAUUUUGUUGUGACUCCAGAGUCACUGGAACCCCAU---------------CCCAGC-CCUUUUCCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAA (((((.............((((((....))))))(((....))).---------------...)))-))......(((...((((((.......)))))).....))).. ( -25.00, z-score = -2.47, R) >droAna3.scaffold_13340 15943546 92 - 23697760 ----AUUGCCAUUUUGUUGUGACUCCAAAGUUACUUC--CUUUAUCC------------UUGUUGCACCACUUCCUGAGUCCUUGCACCUUGAUUGCAAGUUAUUUGCAA ----..............((((((....))))))...--........------------...(((((..((((...)))).((((((.......)))))).....))))) ( -13.50, z-score = -0.66, R) >dp4.chr2 30324300 91 - 30794189 ----AUCGCCAUUUUGUUGUGACUCCAAAGUUACUUG--CCCCGGCCC-----------GUGCUGC-CCCCCUGCCU-CUUCUGGCACCUUGAUUGCAAGUUAUUUGCAA ----.((((.........))))...((((...(((((--(..((((..-----------..)))).-.....((((.-.....))))........))))))..))))... ( -18.90, z-score = -0.85, R) >droPer1.super_6 5715831 91 - 6141320 ----AUCGCCAUUUUGUUGUGACUCCAAAGUUACUUG--CCCCGGCCC-----------GUGCUGC-CCCCCUGCCU-CUUCUGGCACCUUGAUUGCAAGUUAUUUGCAA ----.((((.........))))...((((...(((((--(..((((..-----------..)))).-.....((((.-.....))))........))))))..))))... ( -18.90, z-score = -0.85, R) >droVir3.scaffold_12855 4905564 84 - 10161210 --------ACCCACCAUCUAGGCAAAAGAGUAGAAAAAACUCAG-----------------ACUAU-AGAAGUUUGUAGCCUCUGCACCUUGAUUGCAAGUUAUUUGAAA --------...........((((....((((.......))))((-----------------(((..-...)))))...)))).((((.......))))............ ( -13.00, z-score = 0.13, R) >droMoj3.scaffold_6540 22213706 81 - 34148556 -------GUUGCACUUUGUGGGAGAGAGAGAAGCAGAGGGAGGG-----------------GAAGU-ACAA----GUAACCUCUGCACCUUGAUUGCAAGUUAUUUGAAA -------.(((((((((.....)))).(((..(((((((....(-----------------.....-.)..----....)))))))..)))...)))))........... ( -19.20, z-score = -0.33, R) >consensus ____UUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUCCC____________CCCAGC_CCUUUUGCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAA ..................((((((....))))))...............................................((((((.......)))))).......... ( -7.57 = -8.32 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:39 2011