
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,323,863 – 11,323,957 |
| Length | 94 |
| Max. P | 0.976785 |

| Location | 11,323,863 – 11,323,957 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 69.95 |
| Shannon entropy | 0.53693 |
| G+C content | 0.36357 |
| Mean single sequence MFE | -19.65 |
| Consensus MFE | -10.92 |
| Energy contribution | -13.07 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
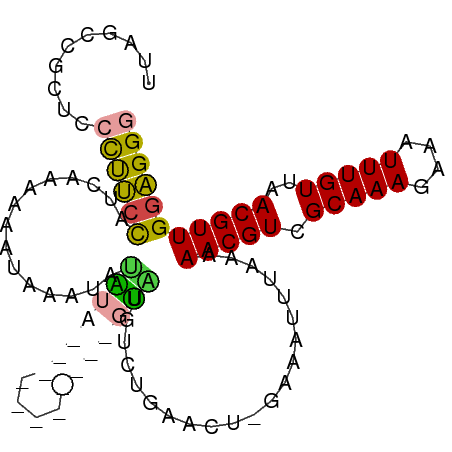
>dm3.chr3R 11323863 94 + 27905053 UUAGCCGCUCCCUUCCAUCAAAAAAUAAAUAUAUA--------UGUGUAUGUCUGAACU-GAAAUUUAAAAACGUCGCAAAGAAAUUUGUUAACGUUGGAGGG ..........((((((.(((........(((((..--------..)))))........)-))........(((((.(((((....)))))..))))))))))) ( -18.79, z-score = -2.08, R) >droSim1.chr3R 17363159 102 - 27517382 UUAGCCGCUCCCUUCCAUCAAAAAAUAAAUAUAUACAUAUAUGUGUGUAUGUCUGAACU-GAAAUUUAAAAACGUCGCAAAGAAAUUUGUUAACGUUGGAGGG ..........((((((.(((........(((((((((....)))))))))........)-))........(((((.(((((....)))))..))))))))))) ( -25.19, z-score = -3.68, R) >droSec1.super_0 10499904 102 - 21120651 UUAGCCGCUCCCUUCCAUCAAAAAAUAAAUAUAUACAUGUGUAUGUGUAUGUCUGAACU-GAAAUUUAAAAACGUCGCAAAGAAAUUUGUUAACGUUGGAGGG ..........((((((............((((((((((....)))))))))).......-..........(((((.(((((....)))))..))))))))))) ( -24.30, z-score = -3.13, R) >droEre2.scaffold_4770 10851350 90 + 17746568 UUAGCCGCUCCCUUCUAUCAGAAUGAAAAUACAA------------GUACGAGAAAACC-GAAAUUUAAAAACGUCGCAAAGAAAUUUGUUAACGUUGGAGGG ....((.(((((((.(((..(.((....)).)..------------))).)))......-..........(((((.(((((....)))))..))))))))))) ( -16.60, z-score = -0.87, R) >droWil1.scaffold_181089 9772247 85 - 12369635 -------UUAGCCUCUCUGGGAGGAUAAAAACGAC-----------GCGGGAGAAAACGAAAAAUUUAAAAACGUCGCAAAGAAAUUUGUUAACGUUGGAGGG -------....(((((((((..(........)..)-----------.)))....................(((((.(((((....)))))..)))))))))). ( -17.60, z-score = -0.99, R) >droVir3.scaffold_12855 4893538 80 + 10161210 ------UUAGCUCCCCAGUAUAUUUACAGUAUGU------------GUGCGUUUGG----GAAAUUUAAAAACGUCGCAAACAAAUUUGUUAACGUUGAGGG- ------....(((((((((((((.........))------------))))...)))----).........(((((.(((((....)))))..))))))))..- ( -15.40, z-score = -0.23, R) >consensus UUAGCCGCUCCCUUCCAUCAAAAAAUAAAUAUAUA___________GUAUGUCUGAACU_GAAAUUUAAAAACGUCGCAAAGAAAUUUGUUAACGUUGGAGGG ..........((((((...............(((((((....))))))).....................(((((.(((((....)))))..))))))))))) (-10.92 = -13.07 + 2.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:38 2011