
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,321,769 – 11,321,898 |
| Length | 129 |
| Max. P | 0.951641 |

| Location | 11,321,769 – 11,321,870 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Shannon entropy | 0.41936 |
| G+C content | 0.34632 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -16.15 |
| Energy contribution | -16.24 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11321769 101 + 27905053 -------------GAAGAUCGCAAACCG-AAUGAAC--CGAAACGACU-UACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUU -------------((..(..((((((..-.......--.....(.(((-(((....)))))).).........(((((((((....))).))))))))))))..)..))......... ( -24.30, z-score = -2.49, R) >droSim1.chr3R 17361009 101 - 27517382 -------------GAAGAUCGCAAACCG-AGCGAAC--CGAAACGACU-UACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUU -------------.....((((......-.))))..--.......(((-(((....))))))((((((((((((((((((((....))).)))))))))))..))))))......... ( -25.40, z-score = -2.53, R) >droSec1.super_0 10497789 101 - 21120651 -------------GAAGAUCGCAAACCG-AGCGAAC--CGAAACGACU-UACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUU -------------.....((((......-.))))..--.......(((-(((....))))))((((((((((((((((((((....))).)))))))))))..))))))......... ( -25.40, z-score = -2.53, R) >droYak2.chr3R 13725585 101 - 28832112 -------------GAAGAUCGCAAAGCG-AAUGAAU--CGAAACGACU-UACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAUUUCGUUUGCUUUUCUCAUUAUAAUU -------------.......(.((((((-((((((.--.(((((.(((-(((....)))))).)....(((.....))).......))))...)))))))))))).)........... ( -25.90, z-score = -2.67, R) >droEre2.scaffold_4770 10849183 101 + 17746568 -------------GAAGAUCGCAAGCCG-AAUGAAC--CGAAACGGCU-UACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUU -------------.....(((.((((((-.......--.....)))))-).))).....(((((((((((((((((((((((....))).)))))))))))..)))))))))...... ( -26.00, z-score = -2.43, R) >droAna3.scaffold_13340 15934098 101 + 23697760 -------------GAUGAUCGCAGUUCG-AAUGAGC--AGGAAAGACC-AACGAAAGUAGGUGGGAAAGAAGCGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUU -------------(((((.....((((.-...))))--.......(((-.((....)).))).(((((((((((((((((((....))).)))))))))).)))))))))))...... ( -27.40, z-score = -2.66, R) >droPer1.super_6 5705970 100 + 6141320 -------------GAAGAUCGC---CCA-AAUAAGCAACGUAUAAAGC-AACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUU -------------.......((---...-.....))............-.((....)).(((((((((((((((((((((((....))).)))))))))))..)))))))))...... ( -18.30, z-score = -1.17, R) >dp4.chr2 30314423 100 + 30794189 -------------GAAGAUCGC---CCA-AAUAAGCAACGUAUAAAGC-AACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUU -------------.......((---...-.....))............-.((....)).(((((((((((((((((((((((....))).)))))))))))..)))))))))...... ( -18.30, z-score = -1.17, R) >droWil1.scaffold_181089 9769111 99 - 12369635 -------------GAAGAUCGUUGCCAA-AAAAAAU--UAAAGCGG---UACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUU -------------....((((((.....-.......--...)))))---)((....)).(((((((((((((((((((((((....))).)))))))))))..)))))))))...... ( -20.86, z-score = -1.85, R) >droVir3.scaffold_12855 4890242 118 + 10161210 GAAGAUCGCUGCAAAACAAAAAAACCAGCAAAAAGGAAAGAAACGCAUGCACGAAUGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUCCCAAUUCGUUUGCUUUUCUCAUUAUAAUU .....(((.((((...........((........))...........))))))).(((((.((((((((((((((((((...........)))))))))))..))))))))))))... ( -21.65, z-score = -0.54, R) >droMoj3.scaffold_6540 22198463 116 + 34148556 GAGGAUCGCUGCCGAAAAUAAGAAAUAA-AAUGAAGCCAAAAAAG-CGGCACGAAUGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUCCCAAUUCGUUUGCUUUUCUCAUUAUAAUU .....(((.(((((..............-................-)))))))).(((((.((((((((((((((((((...........)))))))))))..))))))))))))... ( -23.71, z-score = -0.88, R) >droGri2.scaffold_14906 4525626 117 - 14172833 GAGGAUCGUUCCCCAAAAAAAAAAAAAG-AAGGAACAAAUAAUCGAUGGCACGAAUGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUCCCAAUUCGUUUGCUUUUCUCAUUAUAAUU .......(((((................-..)))))..(((.(((......))).))).((((((((((((((((((((...........)))))))))))..)))))))))...... ( -21.37, z-score = -0.27, R) >consensus _____________GAAGAUCGCAAACCG_AAUGAAC__CGAAACGACU_UACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUU ..................................................((....)).((((((((((((((((((((...........)))))))))))..)))))))))...... (-16.15 = -16.24 + 0.09)


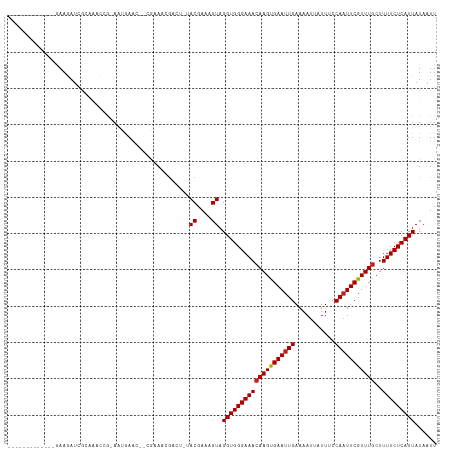
| Location | 11,321,794 – 11,321,892 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.12 |
| Shannon entropy | 0.37349 |
| G+C content | 0.37514 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -16.15 |
| Energy contribution | -16.24 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
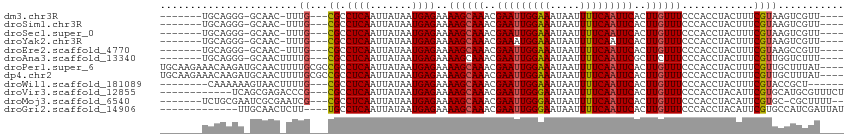
>dm3.chr3R 11321794 98 + 27905053 ----AACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG---CAAA-GUUGC-CCCUGCA------- ----....((((((....))))))(((.(((....(((((((((....))).)))))).(((((...(((((.......))))).)---))))-)))..-)))....------- ( -27.30, z-score = -2.68, R) >droSim1.chr3R 17361034 98 - 27517382 ----AACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG---CAAA-GUUGC-CCCUGCA------- ----....((((((....))))))(((.(((....(((((((((....))).)))))).(((((...(((((.......))))).)---))))-)))..-)))....------- ( -27.30, z-score = -2.68, R) >droSec1.super_0 10497814 98 - 21120651 ----AACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG---CAAA-GUUGC-CCCUGCA------- ----....((((((....))))))(((.(((....(((((((((....))).)))))).(((((...(((((.......))))).)---))))-)))..-)))....------- ( -27.30, z-score = -2.68, R) >droYak2.chr3R 13725610 98 - 28832112 ----AACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAUUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG---CAAA-GUUGC-CCCUGCA------- ----....((((((....))))))(((.(((....(((..((((....))))...))).(((((...(((((.......))))).)---))))-)))..-)))....------- ( -24.70, z-score = -1.79, R) >droEre2.scaffold_4770 10849208 98 + 17746568 ----AACGGCUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG---CAAA-GUUGC-CCCUGCA------- ----....((((((....))))))(((.(((....(((((((((....))).)))))).(((((...(((((.......))))).)---))))-)))..-)))....------- ( -27.60, z-score = -2.25, R) >droAna3.scaffold_13340 15934123 99 + 23697760 ----AAAGACCAACGAAAGUAGGUGGGAAAGAAGCGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG---CAAAAGUUGC-CCCUGCA------- ----....(((.((....)).)))((((((((((((((((((((....))).)))))))))).)))))))...........(((.(---((.....)))-.)))...------- ( -28.60, z-score = -2.83, R) >droPer1.super_6 5705994 110 + 6141320 ----AUAAAGCAACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGGCGCAAAAGUUGCAUCUUGUUUCUUGCA ----........((....))..(..(((((((((.(((((((((....))).))))))..........((((.......))))(((((......)))))..)))))))))..). ( -30.90, z-score = -2.78, R) >dp4.chr2 30314447 110 + 30794189 ----AUAAAGCAACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGGCGCAAAAGUUGCAUCUUGUUUCUUGCA ----........((....))..(..(((((((((.(((((((((....))).))))))..........((((.......))))(((((......)))))..)))))))))..). ( -30.90, z-score = -2.78, R) >droWil1.scaffold_181089 9769136 97 - 12369635 ------AGCGGUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG---CAAAAGUUACUUUUUUG-------- ------.(((.(((....)))(((((((((((((((((((((((....))).)))))))))))..)))))))))..........))---)................-------- ( -23.40, z-score = -2.10, R) >droVir3.scaffold_12855 4890280 99 + 10161210 AGAAACGCAUGCACGAAUGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUCCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG---CGGGUCUCGCUGA------------ (((..(((..((.(((.(((((.((((((((((((((((((...........)))))))))))..)))))))))))).)))..)))---))..)))......------------ ( -26.00, z-score = -1.12, R) >droMoj3.scaffold_6540 22198502 101 + 34148556 --AAAAGCG-GCACGAAUGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUCCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG---CGAUUCGCGAUUCGCAGA------- --....(((-.(.(((.(((((.((((((((((((((((((...........)))))))))))..)))))))))))).))).).))---).....(((...)))...------- ( -27.00, z-score = -1.15, R) >droGri2.scaffold_14906 4525663 97 - 14172833 AUAAUCGAUGGCACGAAUGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUCCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCA----AAGAGUUGCAA------------- ....((..((.(.(((.(((((.((((((((((((((((((...........)))))))))))..)))))))))))).))).).))----..)).......------------- ( -21.30, z-score = -0.98, R) >consensus ____AACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCG___CAAA_GUUGC_CCCUGCA_______ ............((....)).((((((((((((((((((((...........)))))))))))..)))))))))........................................ (-16.15 = -16.24 + 0.09)
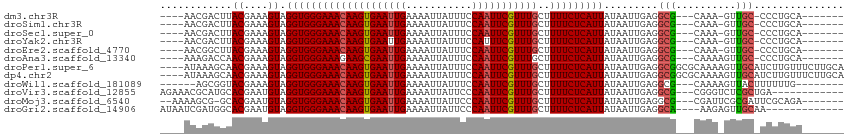
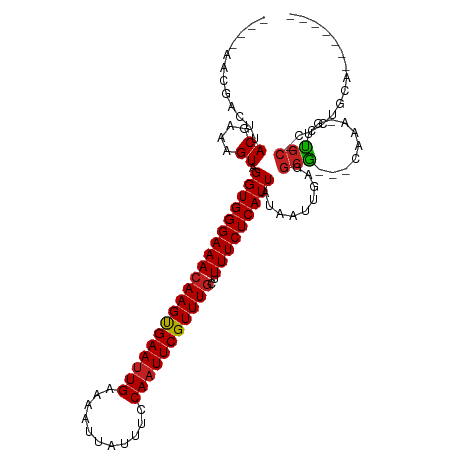
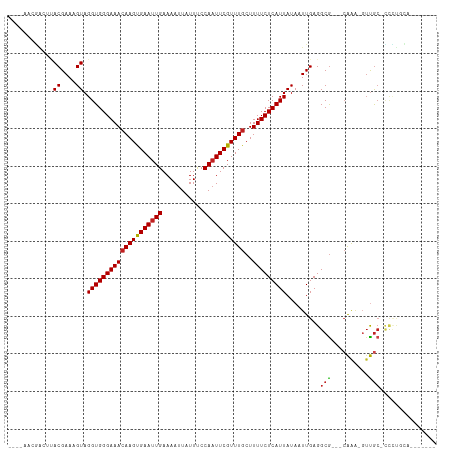
| Location | 11,321,794 – 11,321,892 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.12 |
| Shannon entropy | 0.37349 |
| G+C content | 0.37514 |
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.27 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.812624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11321794 98 - 27905053 -------UGCAGGG-GCAAC-UUUG---CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUU---- -------.((((((-....)-))))---)..((((.......))))..((((((..(((((((((.....)))))))))..))))))...((.(((....))).))....---- ( -22.90, z-score = -2.01, R) >droSim1.chr3R 17361034 98 + 27517382 -------UGCAGGG-GCAAC-UUUG---CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUU---- -------.((((((-....)-))))---)..((((.......))))..((((((..(((((((((.....)))))))))..))))))...((.(((....))).))....---- ( -22.90, z-score = -2.01, R) >droSec1.super_0 10497814 98 + 21120651 -------UGCAGGG-GCAAC-UUUG---CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUU---- -------.((((((-....)-))))---)..((((.......))))..((((((..(((((((((.....)))))))))..))))))...((.(((....))).))....---- ( -22.90, z-score = -2.01, R) >droYak2.chr3R 13725610 98 + 28832112 -------UGCAGGG-GCAAC-UUUG---CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAAUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUU---- -------.((((((-....)-))))---)((((((.......))))....)).(((((...((((((((............))))))))....(((....)))..)))))---- ( -21.00, z-score = -1.41, R) >droEre2.scaffold_4770 10849208 98 - 17746568 -------UGCAGGG-GCAAC-UUUG---CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGCCGUU---- -------.((((((-....)-))))---)((((((.......))))..((((((..(((((((((.....)))))))))..)))))).................))....---- ( -23.50, z-score = -2.04, R) >droAna3.scaffold_13340 15934123 99 - 23697760 -------UGCAGGG-GCAACUUUUG---CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCGCUUCUUUCCCACCUACUUUCGUUGGUCUUU---- -------...((((-((....((((---(..((((.......))))....))))).(((((((((.....))))))))))))))).....(((.((....)).)))....---- ( -23.60, z-score = -1.89, R) >droPer1.super_6 5705994 110 - 6141320 UGCAAGAAACAAGAUGCAACUUUUGCGCCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUUGCUUUAU---- .((((((((((((........(((((.....((((.......))))....))))).(((((((((.....))))))))).)))))))).............)))).....---- ( -24.91, z-score = -3.00, R) >dp4.chr2 30314447 110 - 30794189 UGCAAGAAACAAGAUGCAACUUUUGCGCCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUUGCUUUAU---- .((((((((((((........(((((.....((((.......))))....))))).(((((((((.....))))))))).)))))))).............)))).....---- ( -24.91, z-score = -3.00, R) >droWil1.scaffold_181089 9769136 97 + 12369635 --------CAAAAAAGUAACUUUUG---CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUACCGCU------ --------....((((((...((((---(..((((.......))))....))))).(((((((((.....)))))))))..............)))))).........------ ( -17.50, z-score = -2.55, R) >droVir3.scaffold_12855 4890280 99 - 10161210 ------------UCAGCGAGACCCG---CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGGAAUAAUUUUCAAUUCACUUGUUUCCCACCUACAUUCGUGCAUGCGUUUCU ------------.....((((((.(---(((((((.......))))..((((((..((((((..(.....)..))))))..)))))).............))))..).))))). ( -21.80, z-score = -1.69, R) >droMoj3.scaffold_6540 22198502 101 - 34148556 -------UCUGCGAAUCGCGAAUCG---CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGGAAUAAUUUUCAAUUCACUUGUUUCCCACCUACAUUCGUGC-CGCUUUU-- -------...((((........)))---)..((((.......))))((((((..((((((((((((...................)))))).....))))))..-.))))))-- ( -23.11, z-score = -1.73, R) >droGri2.scaffold_14906 4525663 97 + 14172833 -------------UUGCAACUCUU----UGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGGAAUAAUUUUCAAUUCACUUGUUUCCCACCUACAUUCGUGCCAUCGAUUAU -------------..((......(----(((((((.......))))....))))((((((((((((...................)))))).....)))))))).......... ( -16.01, z-score = -1.11, R) >consensus _______UGCAGGG_GCAAC_UUUG___CGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUU____ ...............................((((.......))))..((((((..(((((((((.....)))))))))..))))))........................... (-12.29 = -12.27 + -0.02)


| Location | 11,321,800 – 11,321,898 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.84 |
| Shannon entropy | 0.38073 |
| G+C content | 0.38649 |
| Mean single sequence MFE | -26.19 |
| Consensus MFE | -16.15 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.886263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
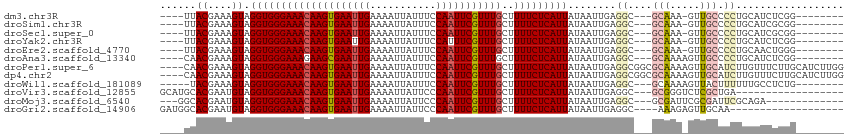
>dm3.chr3R 11321800 98 + 27905053 ----UUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC---GCAAA-GUUGCCCCUGCAUCUCGG-------- ----...(((..(((((.((..(((....(((((((((....))).)))))).(((((...(((((.......))))).---)))))-))).)))))))...))).-------- ( -26.00, z-score = -2.11, R) >droSim1.chr3R 17361040 98 - 27517382 ----UUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC---GCAAA-GUUGCCCCUGCAUCGCGG-------- ----...((...(((((.((..(((....(((((((((....))).)))))).(((((...(((((.......))))).---)))))-))).)))))))....)).-------- ( -25.00, z-score = -1.45, R) >droSec1.super_0 10497820 98 - 21120651 ----UUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC---GCAAA-GUUGCCCCUGCAUCGCGG-------- ----...((...(((((.((..(((....(((((((((....))).)))))).(((((...(((((.......))))).---)))))-))).)))))))....)).-------- ( -25.00, z-score = -1.45, R) >droYak2.chr3R 13725616 98 - 28832112 ----UUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAUUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC---GCAAA-GUUGCCCCUGCAUCUCGG-------- ----...(((..(((((.((..(((....(((..((((....))))...))).(((((...(((((.......))))).---)))))-))).)))))))...))).-------- ( -23.40, z-score = -1.23, R) >droEre2.scaffold_4770 10849214 98 + 17746568 ----UUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC---GCAAA-GUUGCCCCUGCAACUGGG-------- ----.(((....)))(((((((((((((((((((((((....))).)))))))))))..)))))))))....((((((.---(((..-..))).))).))).....-------- ( -25.80, z-score = -1.79, R) >droAna3.scaffold_13340 15934129 99 + 23697760 ----CAACGAAAGUAGGUGGGAAAGAAGCGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC---GCAAAAGUUGCCCCUGCAUCUCGG-------- ----...(((..(((((.((((((((((((((((((((....))).)))))))))).)))))))............(((---((....).)))))))))...))).-------- ( -28.60, z-score = -2.54, R) >droPer1.super_6 5706000 110 + 6141320 ----CAACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGGCGCAAAAGUUGCAUCUUGUUUCUUGCAUCUUGG ----..((....))..(..(((((((((.(((((((((....))).))))))..........((((.......))))(((((......)))))..)))))))))..)....... ( -30.90, z-score = -2.79, R) >dp4.chr2 30314453 110 + 30794189 ----CAACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGGCGCAAAAGUUGCAUCUUGUUUCUUGCAUCUUGG ----..((....))..(..(((((((((.(((((((((....))).))))))..........((((.......))))(((((......)))))..)))))))))..)....... ( -30.90, z-score = -2.79, R) >droWil1.scaffold_181089 9769141 98 - 12369635 -----UACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC---GCAAAAGUUACUUUUUUGCCUCUG-------- -----(((....)))(((((((((((((((((((((((....))).)))))))))))..)))))))))......(((((---(.(((((...))))).))))))..-------- ( -29.80, z-score = -4.32, R) >droVir3.scaffold_12855 4890286 93 + 10161210 GCAUGCACGAAUGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUCCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC---GCGGGUCUCGCUGA------------------ .((.((.....(((((.((((((((((((((((((...........)))))))))))..))))))))))))...(((((---....))))))))).------------------ ( -23.80, z-score = -0.89, R) >droMoj3.scaffold_6540 22198508 95 + 34148556 ---GGCACGAAUGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUCCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC---GCGAUUCGCGAUUCGCAGA------------- ---.((.(((.(((((.((((((((((((((((((...........)))))))))))..)))))))))))).)))..))---((((........))))...------------- ( -25.20, z-score = -1.16, R) >droGri2.scaffold_14906 4525669 91 - 14172833 GAUGGCACGAAUGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUCCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC----AAAGAGUUGCAA------------------- ..((.(.(((.(((((.((((((((((((((((((...........)))))))))))..)))))))))))).))).).)----)...........------------------- ( -19.90, z-score = -0.78, R) >consensus ____UUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGC___GCAAA_GUUGCCCCUGCAUCUCGG________ ......((....)).((((((((((((((((((((...........)))))))))))..)))))))))..............(((.....)))..................... (-16.15 = -16.30 + 0.15)


| Location | 11,321,800 – 11,321,898 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Shannon entropy | 0.38073 |
| G+C content | 0.38649 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.27 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11321800 98 - 27905053 --------CCGAGAUGCAGGGGCAAC-UUUGC---GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA---- --------..(((.(((((((....)-)))))---).))).......((((((.((((((..(((((((((.....)))))))))..)))))).........))))))..---- ( -26.10, z-score = -3.08, R) >droSim1.chr3R 17361040 98 + 27517382 --------CCGCGAUGCAGGGGCAAC-UUUGC---GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA---- --------..((((.((((((....)-)))))---..((((.......))))..((((((..(((((((((.....)))))))))..))))))...........))))..---- ( -25.70, z-score = -2.80, R) >droSec1.super_0 10497820 98 + 21120651 --------CCGCGAUGCAGGGGCAAC-UUUGC---GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA---- --------..((((.((((((....)-)))))---..((((.......))))..((((((..(((((((((.....)))))))))..))))))...........))))..---- ( -25.70, z-score = -2.80, R) >droYak2.chr3R 13725616 98 + 28832112 --------CCGAGAUGCAGGGGCAAC-UUUGC---GCCUCAAUUAUAAUGAGAAAAGCAAACGAAAUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA---- --------..(((.(((((((....)-)))))---).)))....................((((((.((((((((............)))))))).......))))))..---- ( -22.50, z-score = -1.90, R) >droEre2.scaffold_4770 10849214 98 - 17746568 --------CCCAGUUGCAGGGGCAAC-UUUGC---GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA---- --------...(((((.(((.(((..-..)))---.))))))))...((((((.((((((..(((((((((.....)))))))))..)))))).........))))))..---- ( -22.70, z-score = -2.00, R) >droAna3.scaffold_13340 15934129 99 - 23697760 --------CCGAGAUGCAGGGGCAACUUUUGC---GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCGCUUCUUUCCCACCUACUUUCGUUG---- --------.(((((.(.(((((.....(((((---..((((.......))))....)))))((((((((((.....))))))))))........)).))).))))))...---- ( -24.80, z-score = -2.34, R) >droPer1.super_6 5706000 110 - 6141320 CCAAGAUGCAAGAAACAAGAUGCAACUUUUGCGCCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUUG---- ...........((((((((........(((((.....((((.......))))....))))).(((((((((.....))))))))).))))))))................---- ( -24.00, z-score = -2.90, R) >dp4.chr2 30314453 110 - 30794189 CCAAGAUGCAAGAAACAAGAUGCAACUUUUGCGCCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUUG---- ...........((((((((........(((((.....((((.......))))....))))).(((((((((.....))))))))).))))))))................---- ( -24.00, z-score = -2.90, R) >droWil1.scaffold_181089 9769141 98 + 12369635 --------CAGAGGCAAAAAAGUAACUUUUGC---GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUA----- --------..(((((..(((((...)))))..---))))).......((((((.((((((..(((((((((.....)))))))))..)))))).........)))))).----- ( -22.20, z-score = -3.11, R) >droVir3.scaffold_12855 4890286 93 - 10161210 ------------------UCAGCGAGACCCGC---GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGGAAUAAUUUUCAAUUCACUUGUUUCCCACCUACAUUCGUGCAUGC ------------------...(((.....)))---..((((.......))))....(((.((((((((((((...................)))))).....))))))...))) ( -18.61, z-score = -1.18, R) >droMoj3.scaffold_6540 22198508 95 - 34148556 -------------UCUGCGAAUCGCGAAUCGC---GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGGAAUAAUUUUCAAUUCACUUGUUUCCCACCUACAUUCGUGCC--- -------------...((((((((((...)))---).((((.......))))..((((((..((((((..(.....)..))))))..)))))).........))))))...--- ( -20.30, z-score = -1.48, R) >droGri2.scaffold_14906 4525669 91 + 14172833 -------------------UUGCAACUCUUU----GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGGAAUAAUUUUCAAUUCACUUGUUUCCCACCUACAUUCGUGCCAUC -------------------..((......((----((((((.......))))....))))((((((((((((...................)))))).....)))))))).... ( -16.01, z-score = -1.60, R) >consensus ________CCGAGAUGCAGGGGCAAC_UUUGC___GCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA____ .....................................((((.......))))..((((((..(((((((((.....)))))))))..))))))..................... (-12.29 = -12.27 + -0.02)

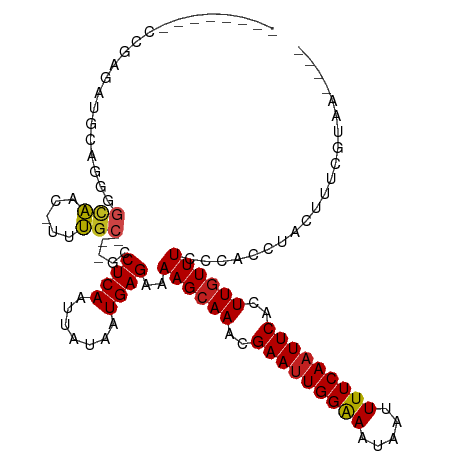

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:37 2011