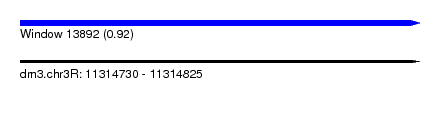
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,314,730 – 11,314,825 |
| Length | 95 |
| Max. P | 0.923883 |

| Location | 11,314,730 – 11,314,825 |
|---|---|
| Length | 95 |
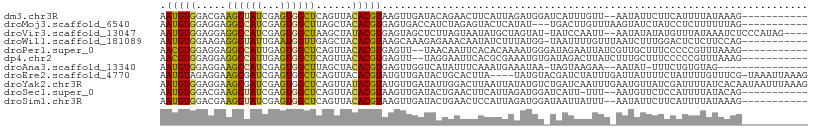
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 59.58 |
| Shannon entropy | 0.86100 |
| G+C content | 0.36718 |
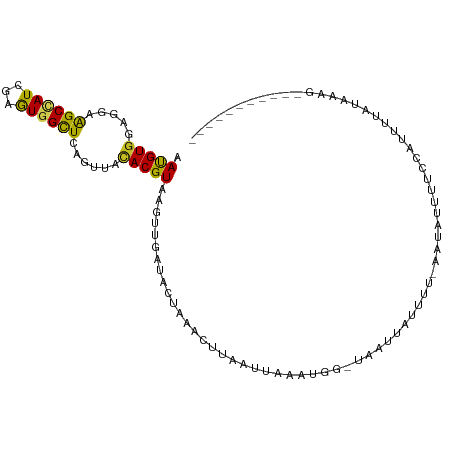
| Mean single sequence MFE | -21.51 |
| Consensus MFE | -9.34 |
| Energy contribution | -8.75 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11314730 95 + 27905053 AAUGUGGACGAAGCUAUCGAGUGGCUCAGUUACACGUAAGUUGAUACAGAACUUCAUUAGAUGGAUCAUUUGUU--AAUAUUCUUCAUUUUAUAAAG----------- ...((((((..((((((...))))))..))).)))(((((.(((...((((......((((((...))))))..--....))))))).)))))....----------- ( -16.10, z-score = 0.25, R) >droMoj3.scaffold_6540 23665928 94 - 34148556 AAUGUGGAGGAGGCCAUCGAGUGGCUUAGCUACACGUGAGUGACCAUCUAGAGUACUCAUAU---UGACUUGUUUAAGUAUCUAUCCUCUUUUUUAG----------- .....((((((((((((...))))))(((.(((..(((((((..(.....)..))))))).(---(((.....))))))).))))))))).......----------- ( -22.90, z-score = -0.46, R) >droVir3.scaffold_13047 5971484 101 - 19223366 AAUGUGGAGGAGGCCAUCGAGUGGCUAAGCUAUACGUGAGUAGCUCUUAGUAAUAUGCUAGUAU-UAUCCAAUU--AAUAUAUAUGUUUAUAAAUCUCCCAUAG---- .(((.(((((.((((((...)))))).((((..((....))))))..((((.....))))((((-((......)--))))).............))))))))..---- ( -23.30, z-score = -0.76, R) >droWil1.scaffold_181089 10858376 96 - 12369635 AAUGUGGAAGAGGCUAUUGAAUGGUUGAGCUACACGUAAGCAAAGAGAAACAAUAUCUUUAUGG-UAAUUUUGUUUAAUCUUUGGACUCUCUUCCAG----------- ....(((((((((...(..((.((((((((......((..((.(((((.......))))).)).-)).....))))))))))..)..))))))))).----------- ( -23.80, z-score = -1.85, R) >droPer1.super_0 11684926 95 - 11822988 AACGUGGAGGAGGCCAUUGAGUGGCUCAGUUACACGUGAGUU--UAACAAUUCACACAAAAUGGGAUAGAAUUAUCGUUGCUUUCCCCCGUUUAAAG----------- ((((.((.((((((((((..(((.....((((.((....)).--))))......)))..)))))((((....)))).....))))))))))).....----------- ( -25.10, z-score = -1.50, R) >dp4.chr2 24480222 95 - 30794189 AACGUGGAGGAGGCCAUUGAGUGGCUCAGUUACACGUGAGUU--UAGGAAUUCACGCGAAAUGUGAUAGACUUAUCUUUGCUUUCCCCCGUUUAAAG----------- ((((.((.(((((.((..((..((.((.((((((((((((((--....)))))))).....)))))).))))..))..)).))))))))))).....----------- ( -27.20, z-score = -1.68, R) >droAna3.scaffold_13340 12902239 89 + 23697760 AAUGUGGAGGAAGCCAUCGAGUGGCUUAGCUACACGUGAGUUGGUCAUAUUUCAAAUGAAAUAA-UAGUAAGAA--AAUAU-UUUCUGUGUAG--------------- ...((((...(((((((...)))))))..))))((....))......((((((....)))))).-.....((((--(....-)))))......--------------- ( -19.80, z-score = -1.60, R) >droEre2.scaffold_4770 10841936 103 + 17746568 AAUGUAGAGGAAGCGAUCGAGUGGCUCAGUUACACGUAUGUUGAUACUGCACUUA----UAUGUACGAUCUAUUUGAUUAUUUUCUAUUUUGUUUCG-UAAAUUAAAG .......((((((..(((((((((.((.((...(((((((.((......))..))----))))))))).)))))))))..))))))...........-.......... ( -20.10, z-score = -0.25, R) >droYak2.chr3R 13718312 108 - 28832112 AAUGUGGAGGAAGCGAUCGAGUGGCUCAGUUAUACGUAUGUUGAUAUUGGACUUAAUUAUAUGUCUGAUCAAUUUGAAUGUUAUCGAUUUUAUCACAAUAAUUUAAAG ..(((((..(((.(((((((((.(.((((....(((((((((((........)))).))))))))))).).))))))......))).)))..)))))........... ( -19.30, z-score = 0.34, R) >droSec1.super_0 10490673 94 - 21120651 AAUGUGGACGAAGCUAUCGAGUGGCUCAGUUACACGUAAGUUGAUACUGAACUUCAUUAGAUGGAUCAUU-UUU--AAUGUUCUCCAUUUUAUACAG----------- ...(((((.(((.(((((.(((((.(((((...((....))....)))))...))))).)))))..(((.-...--.))))))))))).........----------- ( -19.60, z-score = -0.69, R) >droSim1.chr3R 17353924 95 - 27517382 AAUGUGGACGAAGCUAUCGAGUGGCUCAGUUACACGUAAGUUGAUACUGAACUCCAUUAGAUGGAUAAUUAUUU--AAUAUUCUUCAUUUUAUAAAG----------- ..((((((.(((((((((.(((((.(((((...((....))....)))))...))))).))))).(((....))--).....))))..))))))...----------- ( -19.40, z-score = -1.21, R) >consensus AAUGUGGAGGAAGCCAUCGAGUGGCUCAGUUACACGUAAGUUGAUACUAAACUUAAUUAAAUGG_UAAUUAUUUU_AAUAUUUUCCAUUUUAUAAAG___________ .(((((.....((((((...))))))......)))))....................................................................... ( -9.34 = -8.75 + -0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:32 2011