| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,302,251 – 11,302,343 |
| Length | 92 |
| Max. P | 0.947175 |

| Location | 11,302,251 – 11,302,343 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 61.73 |
| Shannon entropy | 0.65069 |
| G+C content | 0.45088 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -12.40 |
| Energy contribution | -13.52 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
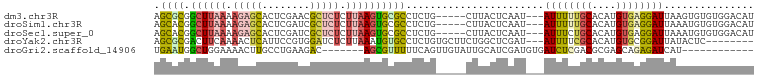
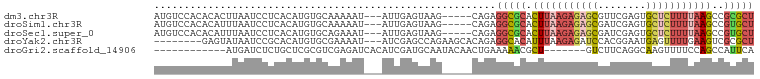
>dm3.chr3R 11302251 92 + 27905053 AGCGCGGCUUAAAAGAGCACUCGAACGCUCUCUUAAGUGCGCCUCUG-----CUUACUCAAU---AUUUUUGCACAUGUGAGGAUUAAGUGUGUGGACAU .(((((.(((((.(((((........))))).)))))))))).((..-----(.((((....---(((((..(....)..)))))..)))).)..))... ( -30.70, z-score = -2.35, R) >droSim1.chr3R 17341537 92 - 27517382 AGCACGGCUUAAAAGAGCACUCGAUCGCUCUCUUAAGUGCGCCUCUG-----CUUACUCAAU---AUUUUUGCACAUGUGAGGAUUAAAUGUGUGGACAU .(((((((((((.(((((........))))).))))))...((((.(-----(.....(((.---....))).....))))))......)))))...... ( -23.10, z-score = -0.67, R) >droSec1.super_0 10473766 92 - 21120651 AGCACGGCUUAAAAGAGCACUCGAUCGCUCUCUUAAGUGCGCCUCUG-----CUUACUCAAU---AUUUCUGCACAUGUGAGGAUUAAAUGUGUGGACAU ((((((((((((.(((((........))))).)))))).))....))-----))........---...((..(((((...........)))))..))... ( -24.40, z-score = -1.03, R) >droYak2.chr3R 13705061 89 - 28832112 AGCGCGACUUCAAAACUCAUUCCGUGGAUCUCUUAAAUGUGCCUCUGUGCUUCUGGCUCGAU---AUUUUCGCACAUGUGCGGAUUAUACUC-------- .((((((..((...((.......)).))..))....((((((....(.((.....)).)((.---....))))))))))))...........-------- ( -17.10, z-score = 0.06, R) >droGri2.scaffold_14906 4424587 81 + 14172833 UGAAUGGCUGGAAAACUUGCCUGAAGAC-------AGCGUUUUUCAGUUGUAUUGCAUCGAUGUGAUCUCGACGCGAGCAGAGAUCAU------------ ...(..(((((((((((((.(....).)-------)).))))))))))..)...........((((((((..(....)..))))))))------------ ( -27.20, z-score = -2.31, R) >consensus AGCACGGCUUAAAAGAGCACUCGAUCGCUCUCUUAAGUGCGCCUCUG_____CUUACUCAAU___AUUUUUGCACAUGUGAGGAUUAAAUGUGUGGACAU .((((.((((((.(((((........))))).)))))))))).......................((((((((....))))))))............... (-12.40 = -13.52 + 1.12)
| Location | 11,302,251 – 11,302,343 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 61.73 |
| Shannon entropy | 0.65069 |
| G+C content | 0.45088 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -9.76 |
| Energy contribution | -11.16 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
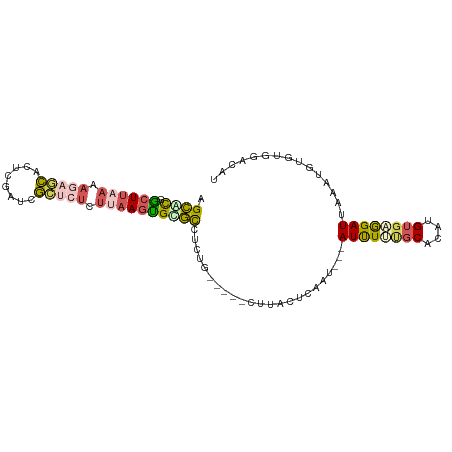
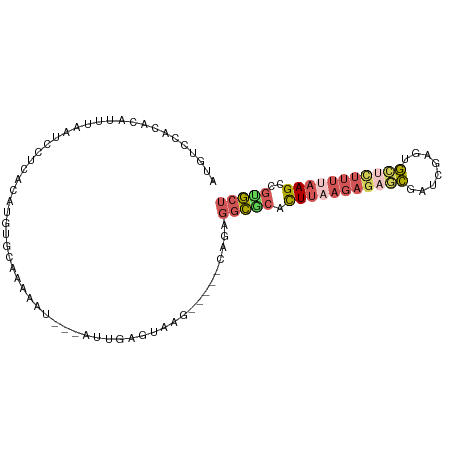
>dm3.chr3R 11302251 92 - 27905053 AUGUCCACACACUUAAUCCUCACAUGUGCAAAAAU---AUUGAGUAAG-----CAGAGGCGCACUUAAGAGAGCGUUCGAGUGCUCUUUUAAGCCGCGCU ...........((((...((((.((((......))---))))))))))-----....(((((.((((((((((((......))))))))))))..))))) ( -27.60, z-score = -2.19, R) >droSim1.chr3R 17341537 92 + 27517382 AUGUCCACACAUUUAAUCCUCACAUGUGCAAAAAU---AUUGAGUAAG-----CAGAGGCGCACUUAAGAGAGCGAUCGAGUGCUCUUUUAAGCCGUGCU ..................((((.((((......))---))))))....-----....(((((.((((((((((((......))))))))))))..))))) ( -25.00, z-score = -1.69, R) >droSec1.super_0 10473766 92 + 21120651 AUGUCCACACAUUUAAUCCUCACAUGUGCAGAAAU---AUUGAGUAAG-----CAGAGGCGCACUUAAGAGAGCGAUCGAGUGCUCUUUUAAGCCGUGCU ..................((((.((((......))---))))))....-----....(((((.((((((((((((......))))))))))))..))))) ( -25.00, z-score = -1.38, R) >droYak2.chr3R 13705061 89 + 28832112 --------GAGUAUAAUCCGCACAUGUGCGAAAAU---AUCGAGCCAGAAGCACAGAGGCACAUUUAAGAGAUCCACGGAAUGAGUUUUGAAGUCGCGCU --------.................((((((....---.(((((((...........))).(((((..(.....)...)))))....))))..)))))). ( -14.80, z-score = 1.07, R) >droGri2.scaffold_14906 4424587 81 - 14172833 ------------AUGAUCUCUGCUCGCGUCGAGAUCACAUCGAUGCAAUACAACUGAAAAACGCU-------GUCUUCAGGCAAGUUUUCCAGCCAUUCA ------------.............(((((((.......))))))).......(((.(((((..(-------(((....)))).))))).)))....... ( -20.30, z-score = -1.48, R) >consensus AUGUCCACACAUUUAAUCCUCACAUGUGCAAAAAU___AUUGAGUAAG_____CAGAGGCGCACUUAAGAGAGCGAUCGAGUGCUCUUUUAAGCCGUGCU .........................................................(((((.(((((((((((........)))))))))))..))))) ( -9.76 = -11.16 + 1.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:30 2011