| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,298,058 – 11,298,177 |
| Length | 119 |
| Max. P | 0.738614 |

| Location | 11,298,058 – 11,298,164 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 64.81 |
| Shannon entropy | 0.73693 |
| G+C content | 0.43502 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -9.24 |
| Energy contribution | -9.54 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.23 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.738614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
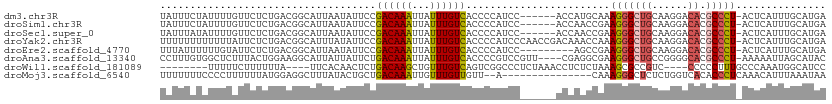
>dm3.chr3R 11298058 106 + 27905053 -UUACUGCCUGUGGUUUAUUUCUAUUUUGU--UCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC------ACCAUGCAAAGGGCUGCAAGGACACGCCCUA -....(((.((.((..((((...((.((((--(....))))).)).))))..))(((((.....))))).........------..)).))).(((((((......)).))))). ( -21.00, z-score = -0.23, R) >droSim1.chr3R 17337288 107 - 27517382 GUUACUGCCUGUGGUUUAUUUCUAUUUUGU--UCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC------ACCAACCGAAGGGCUGCAAGGACACGCCCUA ((((.((((.((((.......))))...((--(....))))))).)))).....(((((.....))))).........------.........(((((((......)).))))). ( -20.30, z-score = 0.02, R) >droSec1.super_0 10469545 107 - 21120651 GUUACUGCCUGUAGUUUAUUUAUAUUUUGU--UCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC------ACCAACCGAAGGGCUGCAAGGACACGCCCUA ((((.(((((((((.....)))))....((--(....))))))).)))).....(((((.....))))).........------.........(((((((......)).))))). ( -20.40, z-score = -0.57, R) >droYak2.chr3R 13700490 113 - 28832112 GUUACUGCUUGUGGCUUUUUUUUUUUUUAU--UCUCUGACGGCAUUUAUAUUCCGACAAAUUAUUUGUCACCCCAUCCCAACCGACAAACCAAAGGGCUGCAAGGACACGCCCUA .......((((..(((((((..........--....((.(((.((....)).))).)).....((((((..............))))))..)))))))..))))........... ( -20.74, z-score = -0.47, R) >droEre2.scaffold_4770 10824813 106 + 17746568 GUAACUGCUUGUGAUUAUUUUAUUUUUUGUAUUCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC------AGC---CGAAGGGCUGCAAGGACACGCCCUA ((....(..((..(..(......)..)..))..)....))(((...........(((((.....)))))...((...(------(((---(....)))))...))....)))... ( -21.90, z-score = -0.87, R) >droAna3.scaffold_13340 12885082 109 + 23697760 GUUAUUGUUUCGCUUGCCUUUGUGGCUCUU--UACUGGAAGGCAUUAUUAUUCUGACAAAUUAUUUGUCACCCCGUCC----GUUCGAGGCGAAGGGCUGCCGGGGCACGCCCUA ...........((.(((((..(..((((((--(.((.(((((.((........((((((.....))))))....))))----.))).))..)))))))..)..))))).)).... ( -34.60, z-score = -1.30, R) >droWil1.scaffold_181089 10837137 97 - 12369635 -------GUCAGAGUUUUUUCUUUUUUUCU--UUUUUA-----UUCACAACUCUGACAAGCUGUUUGUCAGUCGGCCCUCUAAACCUCUCUAAAGCGCCGUC----CCCCCUUUG -------(((((((((..............--......-----.....)))))))))..((((.....))))((((.((.((........)).)).))))..----......... ( -19.11, z-score = -3.68, R) >droMoj3.scaffold_6540 23647045 82 - 34148556 --------------UUUUUUUUUCCCCUUU--UUUAUGGAGGCUUUAUACUGCUGACAAAUUGUUUGUUGUUACA-----------------AAGGGCUCUCUGGUCACACCCUC --------------..........(((...--....((..(((........)))..)).....(((((....)))-----------------)))))......((.....))... ( -11.80, z-score = 0.89, R) >consensus GUUACUGCUUGUGGUUUUUUUUUAUUUUGU__UCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC______ACCAACCGAAGGGCUGCAAGGACACGCCCUA ......................................................((((((...))))))........................(((((((......)).))))). ( -9.24 = -9.54 + 0.30)


| Location | 11,298,073 – 11,298,177 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.34 |
| Shannon entropy | 0.68954 |
| G+C content | 0.42862 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -8.71 |
| Energy contribution | -9.01 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.23 |
| Mean z-score | -0.62 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11298073 104 + 27905053 UAUUUCUAUUUUGUUCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC------ACCAUGCAAAGGGCUGCAAGGACACGCCCU-ACUCAUUUGCAUGA ......................((.((....)).))(((((.....))))).........------..((((((((((((((......)).)))))-......))))))). ( -20.70, z-score = -1.09, R) >droSim1.chr3R 17337304 104 - 27517382 UAUUUCUAUUUUGUUCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC------ACCAACCGAAGGGCUGCAAGGACACGCCCU-ACUCAUUUGCAUGA ...........((((((.((.((((...........(((((.....)))))...((.((.------.......)).))))))))))))))......-.............. ( -18.00, z-score = -0.38, R) >droSec1.super_0 10469561 104 - 21120651 UAUUUAUAUUUUGUUCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC------ACCAACCGAAGGGCUGCAAGGACACGCCCU-ACUCAUUUGCAUGA ...........((((((.((.((((...........(((((.....)))))...((.((.------.......)).))))))))))))))......-.............. ( -18.00, z-score = -0.38, R) >droYak2.chr3R 13700506 110 - 28832112 UUUUUUUUUUUUAUUCUCUGACGGCAUUUAUAUUCCGACAAAUUAUUUGUCACCCCAUCCCAACCGACAAACCAAAGGGCUGCAAGGACACGCCCU-ACUCAUUUGCAUGA .......................(((..........(((((.....)))))........................(((((((......)).)))))-.......))).... ( -15.90, z-score = -0.08, R) >droEre2.scaffold_4770 10824831 101 + 17746568 UUUAUUUUUUGUAUUCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC---------AGCCGAAGGGCUGCAAGGACACGCCCU-ACUCAUUUGCAUGA .........((((.........(((...........(((((.....)))))...((...(---------((((....)))))...))....)))..-.......))))... ( -23.27, z-score = -1.90, R) >droAna3.scaffold_13340 12885098 106 + 23697760 CCUUUGUGGCUCUUUACUGGAAGGCAUUAUUAUUCUGACAAAUUAUUUGUCACCCCGUCCGUU----CGAGGCGAAGGGCUGCCGGGGCACGCCCU-AAAAAUUAGCAUAC .....(..(((((((.((.(((((.((........((((((.....))))))....)))).))----).))..)))))))..).((((....))))-.............. ( -27.40, z-score = 0.12, R) >droWil1.scaffold_181089 10837153 95 - 12369635 --------UUUUUCUUUUUUA----UUCACAACUCUGACAAGCUGUUUGUCAGUCGGCCCUCUAAACCUCUCUAAAGCGCCGUC----CCCCCUUUGCCCAAAUGGCAUCC --------.............----.........((((((((...)))))))).((((.((.((........)).)).))))..----.......((((.....))))... ( -17.10, z-score = -2.27, R) >droMoj3.scaffold_6540 23647047 94 - 34148556 UUUUUUUCCCCUUUUUUAUGGAGGCUUUAUACUGCUGACAAAUUGUUUGUUGUU--A---------------CAAAGGGCUCUCUGGUCACACCCUCAAACAUUUAAAUAA ..................((..(((........)))..))..(((((((.((((--.---------------...((((.....((....))))))..))))..))))))) ( -12.60, z-score = 1.03, R) >consensus UAUUUUUAUUUUGUUCUCUGACGGCAUUAAUAUUCCGACAAAUUAUUUGUCACCCCAUCC______ACCAACCAAAGGGCUGCAAGGACACGCCCU_ACUCAUUUGCAUGA ....................................((((((...))))))........................(((((((......)).)))))............... ( -8.71 = -9.01 + 0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:28 2011