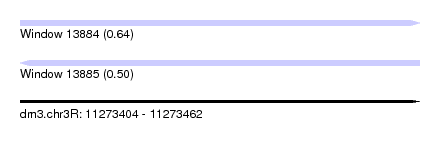
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,273,404 – 11,273,462 |
| Length | 58 |
| Max. P | 0.641081 |

| Location | 11,273,404 – 11,273,462 |
|---|---|
| Length | 58 |
| Sequences | 14 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 71.86 |
| Shannon entropy | 0.65702 |
| G+C content | 0.54698 |
| Mean single sequence MFE | -16.43 |
| Consensus MFE | -8.08 |
| Energy contribution | -7.94 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.641081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11273404 58 + 27905053 AGUCAUUGCCGAAACUCAGCUGGCACUGGGCGUCCAGGGAGUAGAUCUCUCCAGGCAG ......((((((..(((((......)))))..))...((((.......)))).)))). ( -18.10, z-score = -0.28, R) >droSim1.chr3R 17312153 58 - 27517382 AGUCAUUGCCGAAACUCAGCUGGCACUGGGCGUCCAGGGAGUAAAUCUCUCCAGGCAG ......((((((..(((((......)))))..))...((((.......)))).)))). ( -18.10, z-score = -0.82, R) >droSec1.super_0 10445255 58 - 21120651 AGUCAUUGCCGAAACUCAGCUGGCACUGGGCGUCCAGGGAGUAUAUCUCUCCAGGCAG ......((((((..(((((......)))))..))...((((.......)))).)))). ( -18.10, z-score = -0.77, R) >droYak2.chr3R 13674901 58 - 28832112 AGUCGUUGCCGAAACUCAGCUGGCACUGGGCGUCCAGGGAGUAGAUCUCUCCGGGCAG .(((..(((((.........)))))...)))((((.(((((.....))))).)))).. ( -20.10, z-score = -0.41, R) >droEre2.scaffold_4770 10800232 58 + 17746568 AGUCAUUGCCGAAACUCAGCUGGCACUGGGCGUCCAGGGAGUAGAUCUCCCCGGGCAG .(((..(((((.........)))))...)))((((.(((((.....))))).)))).. ( -24.20, z-score = -1.86, R) >droAna3.scaffold_13340 12859268 58 + 23697760 UCGAGUUGCCGAAAAUCAACUGGCACUGGUGGGUUAGGGAGUAGAUCUCGCCAGGCAG ...(((((........))))).((.((((((((((........)))).)))))))).. ( -17.40, z-score = -0.94, R) >dp4.chr2 24439716 58 - 30794189 UCUGGUUGCCGUAAAUCAGCUCGCACUGGUUGUCCAGCGAGUAGAUCUCGCCCGGCAG ......(((((...(((.((((((..(((....))))))))).)))......))))). ( -20.30, z-score = -1.91, R) >droPer1.super_0 11644380 58 - 11822988 UCUGGUUGCCGUAAAUCAGCUCGCACUGGUUGUCCAGCGAGUAGAUCUCGCCCGGCAG ......(((((...(((.((((((..(((....))))))))).)))......))))). ( -20.30, z-score = -1.91, R) >droWil1.scaffold_181089 10809720 55 - 12369635 ---UAUUACUGUAUAUCAAUUGACAUUGGCUAUCAAGGGAGUAGAUUUCACCCGGUAG ---...(((((....(((((....))))).......(((((.....))).))))))). ( -6.50, z-score = 0.97, R) >droVir3.scaffold_13047 5919055 58 - 19223366 UCUGGUUGCCAUGUAUCAGCUGGCAUUGGUGGUCCAGCGAGUAAAUCUCACCGGGCAG .(..((.((((.((....))))))))..)..((((.(.(((.....))).).)))).. ( -17.80, z-score = -0.62, R) >droMoj3.scaffold_6540 23614456 58 - 34148556 UCUGGUUGCCAUGUAUCAGACGACACUGAUGGUCCAGCGAAUAAAUCUCUCCUGGCUG .((((..((((....((((......))))))))))))..................... ( -10.30, z-score = 0.68, R) >droGri2.scaffold_14906 4390836 58 + 14172833 UCUGGUUGCCAUGUAUCAGCUGACACUGGUGAUCCAGAGAGUAGAUCUCACCGGGCAG ((((((..(((.((.((....)).)))))..).)))))(((.....)))......... ( -17.60, z-score = -0.46, R) >anoGam1.chr2R 53520604 58 + 62725911 AUCCAUUGCCAAACACCAGCUCGCACUGCUUGUCGUUGGUGAACUUUUCUCCCGCCAG .....(..((((..((.(((.......))).))..))))..)................ ( -11.00, z-score = -0.87, R) >triCas2.ChLG6 4155970 58 + 13544221 AACCAAUCCCGAAUUCUAGUUCGCAUUGUUUGUCGGCUUCGAAGUUCUCUCCUGGUAG .((((.....(((((..(((..(((.....)))..)))....))))).....)))).. ( -10.20, z-score = -0.91, R) >consensus ACUCAUUGCCGAAAAUCAGCUGGCACUGGUCGUCCAGGGAGUAGAUCUCUCCAGGCAG ......((((....(((((......)))))........(((.....)))....)))). ( -8.08 = -7.94 + -0.14)

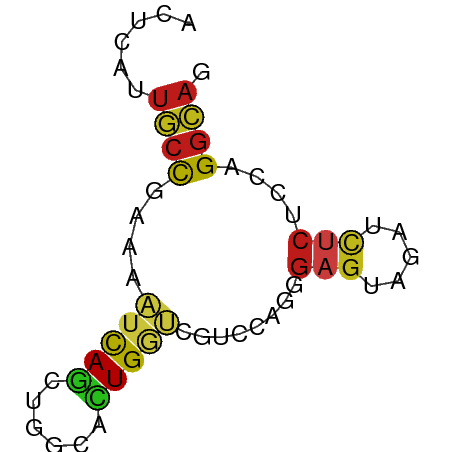
| Location | 11,273,404 – 11,273,462 |
|---|---|
| Length | 58 |
| Sequences | 14 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 71.86 |
| Shannon entropy | 0.65702 |
| G+C content | 0.54698 |
| Mean single sequence MFE | -16.09 |
| Consensus MFE | -6.79 |
| Energy contribution | -7.42 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 11273404 58 - 27905053 CUGCCUGGAGAGAUCUACUCCCUGGACGCCCAGUGCCAGCUGAGUUUCGGCAAUGACU .((((.((.(((.....)))))..((.((.((((....)))).)).))))))...... ( -16.60, z-score = -0.34, R) >droSim1.chr3R 17312153 58 + 27517382 CUGCCUGGAGAGAUUUACUCCCUGGACGCCCAGUGCCAGCUGAGUUUCGGCAAUGACU .((((.((.(((.....)))))..((.((.((((....)))).)).))))))...... ( -16.60, z-score = -0.72, R) >droSec1.super_0 10445255 58 + 21120651 CUGCCUGGAGAGAUAUACUCCCUGGACGCCCAGUGCCAGCUGAGUUUCGGCAAUGACU .((((.((.(((.....)))))..((.((.((((....)))).)).))))))...... ( -16.60, z-score = -0.81, R) >droYak2.chr3R 13674901 58 + 28832112 CUGCCCGGAGAGAUCUACUCCCUGGACGCCCAGUGCCAGCUGAGUUUCGGCAACGACU .(((((((.(((.....))).)))((.((.((((....)))).)).))))))...... ( -17.00, z-score = -0.47, R) >droEre2.scaffold_4770 10800232 58 - 17746568 CUGCCCGGGGAGAUCUACUCCCUGGACGCCCAGUGCCAGCUGAGUUUCGGCAAUGACU .(((((((((((.....)))))))((.((.((((....)))).)).))))))...... ( -23.90, z-score = -2.17, R) >droAna3.scaffold_13340 12859268 58 - 23697760 CUGCCUGGCGAGAUCUACUCCCUAACCCACCAGUGCCAGUUGAUUUUCGGCAACUCGA .(((((((((((.....)))....((......))))))...(.....)))))...... ( -10.00, z-score = 0.36, R) >dp4.chr2 24439716 58 + 30794189 CUGCCGGGCGAGAUCUACUCGCUGGACAACCAGUGCGAGCUGAUUUACGGCAACCAGA .(((((....(((((..((((((((....)))..)))))..))))).)))))...... ( -22.80, z-score = -3.00, R) >droPer1.super_0 11644380 58 + 11822988 CUGCCGGGCGAGAUCUACUCGCUGGACAACCAGUGCGAGCUGAUUUACGGCAACCAGA .(((((....(((((..((((((((....)))..)))))..))))).)))))...... ( -22.80, z-score = -3.00, R) >droWil1.scaffold_181089 10809720 55 + 12369635 CUACCGGGUGAAAUCUACUCCCUUGAUAGCCAAUGUCAAUUGAUAUACAGUAAUA--- .(((.(((.(.......).)))((((((.....))))))..........)))...--- ( -7.40, z-score = 0.09, R) >droVir3.scaffold_13047 5919055 58 + 19223366 CUGCCCGGUGAGAUUUACUCGCUGGACCACCAAUGCCAGCUGAUACAUGGCAACCAGA (((.((((((((.....))))))))........(((((.........)))))..))). ( -19.70, z-score = -2.88, R) >droMoj3.scaffold_6540 23614456 58 + 34148556 CAGCCAGGAGAGAUUUAUUCGCUGGACCAUCAGUGUCGUCUGAUACAUGGCAACCAGA ..((((..(.((((.....(((((......)))))..))))..)...))))....... ( -13.00, z-score = -0.08, R) >droGri2.scaffold_14906 4390836 58 - 14172833 CUGCCCGGUGAGAUCUACUCUCUGGAUCACCAGUGUCAGCUGAUACAUGGCAACCAGA .((((.(((((..((((.....))))))))).(((((....)))))..))))...... ( -18.80, z-score = -1.28, R) >anoGam1.chr2R 53520604 58 - 62725911 CUGGCGGGAGAAAAGUUCACCAACGACAAGCAGUGCGAGCUGGUGUUUGGCAAUGGAU ...((.(((.(..(((((....((........))..)))))..).))).))....... ( -11.60, z-score = 0.88, R) >triCas2.ChLG6 4155970 58 - 13544221 CUACCAGGAGAGAACUUCGAAGCCGACAAACAAUGCGAACUAGAAUUCGGGAUUGGUU ..(((((....(((.(((.....((.((.....)))).....))))))....))))). ( -8.50, z-score = 0.36, R) >consensus CUGCCCGGAGAGAUCUACUCCCUGGACAACCAGUGCCAGCUGAUUUUCGGCAACGAGU .(((((((.(((.....))).)))......((((....))))......))))...... ( -6.79 = -7.42 + 0.63)
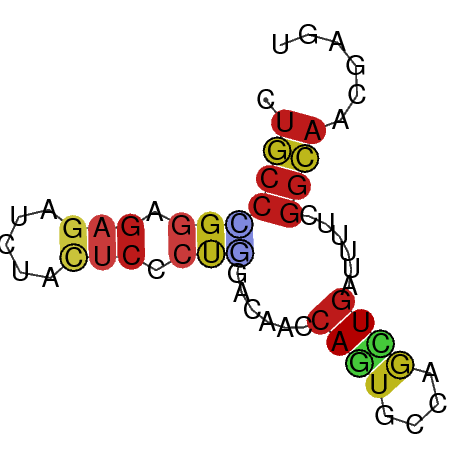
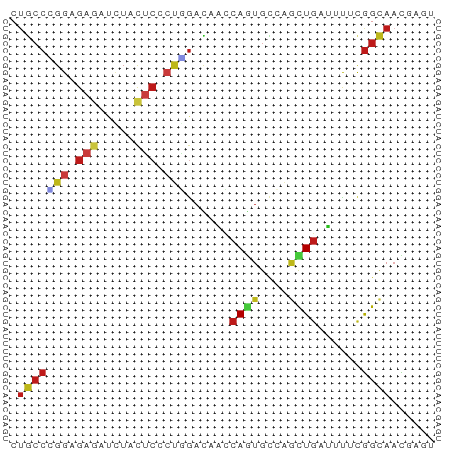
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:27 2011