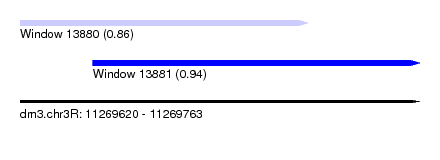
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,269,620 – 11,269,763 |
| Length | 143 |
| Max. P | 0.940214 |

| Location | 11,269,620 – 11,269,723 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.47412 |
| G+C content | 0.57449 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -12.80 |
| Energy contribution | -14.22 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
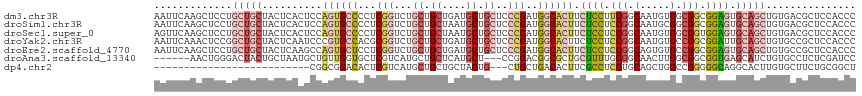
>dm3.chr3R 11269620 103 + 27905053 UCCGCGAUUCCGUGAGCCUCCGAUUGAAUUCAAGCUCCUGCUGCUACUCACUCCAGUGCCCCUCGGUCUGCUGCUGAUGCUGCUCCCGAUGGCACUUCUCCUU ...........(((((.....((......)).(((....)))....)))))...((((((..((((...((.((....)).))..)))).))))))....... ( -28.40, z-score = -1.84, R) >droSim1.chr3R 17308419 103 - 27517382 UCCGCGAUUCCGUGAGCCUCCAAUUGAAUUCAAGCUCCUGCUGCUACUCACUCCAGUGCCCCUCGGUCUGCUGCUAAUGCUGCUCCCGAUGGCACUUCUCCUC ...........(((((........((....))(((....)))....)))))...((((((..((((...((.((....)).))..)))).))))))....... ( -27.40, z-score = -2.43, R) >droSec1.super_0 10441459 103 - 21120651 ACCGCGAUUCCGUGAGCCUCCAAUUGAGUUCAAGCUCCUGCUGCUACUCACUCCAGUGCCCCUCGGUCUGCUGCUAAUGCUGCUCCCGAUGGCACUUCUCCUC ..((((....))))..........(((((...(((....)))...)))))....((((((..((((...((.((....)).))..)))).))))))....... ( -32.20, z-score = -3.40, R) >droYak2.chr3R 13671178 103 - 28832112 UCCCCGAUUCCGUAAGCCACCAAUUGAAUUCAAACUCCGGCUGCUACUCAAUCCCGUGCCACGCGGUCUGCUGCUGAUGCUGCUCCCGAUGGCACUUCUCCUC .....((((..(((((((.....(((....))).....))))..)))..))))..((((((((.((...((.((....)).)).)))).))))))........ ( -27.90, z-score = -2.86, R) >droEre2.scaffold_4770 10796218 103 + 17746568 UCCGCGAUGCCGUACACUUCCAAUUGAAUUCAAGCUCCUGCUGCUACUCAAGCCAGUGCUCCUCGGUCUGCUGCUGAUGCUGCUCCCGAUGGCACUUCUCCUC .......((((((.((((.....((((.....(((....))).....))))...)))).....(((...((.((....)).))..)))))))))......... ( -24.20, z-score = -0.49, R) >droAna3.scaffold_13340 12855468 82 + 23697760 ----------------CCUUUGGGUC--CUUGAACUGGGACUACUGCUAAUGCUGUUGGUGCUCGUCAUGCUGCUCAUGC---UCCGGACGGCGCUGCGUUUG ----------------......((((--((......))))))......(((((....(((((.((((.((..((....))---..)))))))))))))))).. ( -26.00, z-score = -1.25, R) >consensus UCCGCGAUUCCGUGAGCCUCCAAUUGAAUUCAAGCUCCUGCUGCUACUCACUCCAGUGCCCCUCGGUCUGCUGCUGAUGCUGCUCCCGAUGGCACUUCUCCUC ........................(((.(...(((....)))...).)))....((((((..((((...((.((....)).))..)))).))))))....... (-12.80 = -14.22 + 1.42)


| Location | 11,269,646 – 11,269,763 |
|---|---|
| Length | 117 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.36 |
| Shannon entropy | 0.56157 |
| G+C content | 0.62458 |
| Mean single sequence MFE | -42.59 |
| Consensus MFE | -20.31 |
| Energy contribution | -22.54 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11269646 117 + 27905053 AAUUCAAGCUCCUGCUGCUACUCACUCCAGUGCCCCUCGGUCUGCUGCUGAUGCUGCUCCCGAUGGCACUUCUCCUUCGGCAAUGUGGCGGCGGAGUGCAGCUGUGACGCUCCACCC .............((((((((.......((((((..((((...((.((....)).))..)))).))))))....(....)....))))))))((((((((....)).)))))).... ( -43.10, z-score = -1.78, R) >droSim1.chr3R 17308445 117 - 27517382 AAUUCAAGCUCCUGCUGCUACUCACUCCAGUGCCCCUCGGUCUGCUGCUAAUGCUGCUCCCGAUGGCACUUCUCCUCCGGCAAUGCGGCGGCGGAGUGCAGCUGUGACGCUCCACCC ......((((((.(((((..........((((((..((((...((.((....)).))..)))).)))))).((((.(((.(.....).))).)))).))))).).)).)))...... ( -44.30, z-score = -2.36, R) >droSec1.super_0 10441485 117 - 21120651 AGUUCAAGCUCCUGCUGCUACUCACUCCAGUGCCCCUCGGUCUGCUGCUAAUGCUGCUCCCGAUGGCACUUCUCCUCCGGCAAUGUGGCGGUGGAGUGCAGCUGUGACGCUCCACCC ......((((((.(((((..........((((((..((((...((.((....)).))..)))).)))))).((((.(((.(.....).))).)))).))))).).)).)))...... ( -44.90, z-score = -2.24, R) >droYak2.chr3R 13671204 117 - 28832112 AAUUCAAACUCCGGCUGCUACUCAAUCCCGUGCCACGCGGUCUGCUGCUGAUGCUGCUCCCGAUGGCACUUCUCCUCCGGCAAUGUGCCGGCGGAUUGCAGCUGUGCCGCUCCACCC ...........(((((((...........((((((((.((...((.((....)).)).)))).))))))...(((.(((((.....))))).)))..)))))))............. ( -46.20, z-score = -3.58, R) >droEre2.scaffold_4770 10796244 117 + 17746568 AAUUCAAGCUCCUGCUGCUACUCAAGCCAGUGCUCCUCGGUCUGCUGCUGAUGCUGCUCCCGAUGGCACUUCUCCUCCGGCAGUGUGCCGGCGGAGUGCAGCUGUGCCGCUCCACCC .......((.(..(((((..........((((((..((((...((.((....)).))..)))).)))))).((((.(((((.....))))).)))).))))).).)).......... ( -45.50, z-score = -1.71, R) >droAna3.scaffold_13340 12855482 108 + 23697760 ------AACUGGGACUACUGCUAAUGCUGUUGGUGCUCGUCAUGCUGCUCAUGCU---CCGGACGGCGCUGCGUUUGCGGCAACUUGGCGGCGGUGAGCAUCUGUGCCUCUCGAUCC ------....((.((...((((..((((((..(.((.((((.((..((....)).---.))))))))(((((....)))))...)..))))))...))))...)).))......... ( -39.70, z-score = -0.69, R) >dp4.chr2 24435492 88 - 30794189 --------------------------CGGCGGACACUCGUCAUGCUGCUGCUACUG---CUGCUGACACUUCGCCUCCUGCAGCUGGCCGGGGGCAGGCACUUGUGCUUCUGCGGCU --------------------------((((((((....))).)))))........(---((((.((......((((((.((.....)).)))))).((((....)))))).))))). ( -34.40, z-score = 0.06, R) >consensus AAUUCAAGCUCCUGCUGCUACUCACUCCAGUGCCCCUCGGUCUGCUGCUGAUGCUGCUCCCGAUGGCACUUCUCCUCCGGCAAUGUGGCGGCGGAGUGCAGCUGUGCCGCUCCACCC .............(((((..........((((((...(((...((.((....)).))..)))..)))))).((((.(((.(.....).))).)))).)))))............... (-20.31 = -22.54 + 2.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:24 2011