| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,255,888 – 11,255,996 |
| Length | 108 |
| Max. P | 0.913436 |

| Location | 11,255,888 – 11,255,996 |
|---|---|
| Length | 108 |
| Sequences | 8 |
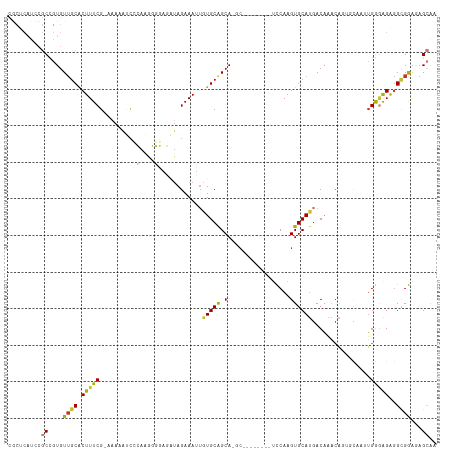
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.80 |
| Shannon entropy | 0.58626 |
| G+C content | 0.49222 |
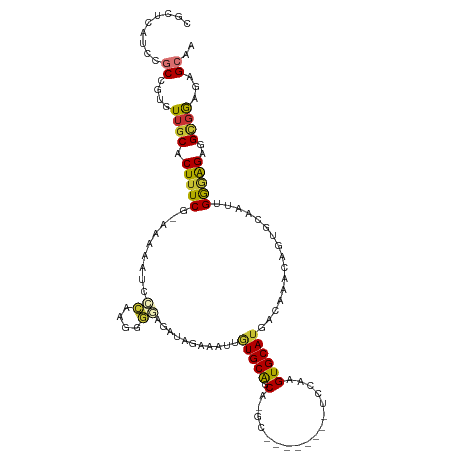
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -7.44 |
| Energy contribution | -7.30 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
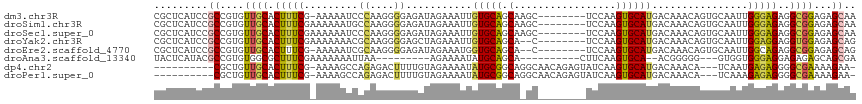
>dm3.chr3R 11255888 108 - 27905053 CGCUCAUCCGCCGUGUUGCACUUUCG-AAAAAUCCCAAGGGGAGAUAGAAAUUGUGCAGCAAGC--------UCCAAGUGCAUGACAAACAGUGCAAUUGGGAGAGGCGGAGAGCAA .((((.((((((.((((((((((((.-.....((((...))))....))))..))))))))..(--------(((...(((((........)))))....)))).)))))))))).. ( -50.10, z-score = -6.12, R) >droSim1.chr3R 17293104 109 + 27517382 CGCUCAUCCGCCGUGUUGCACUUUCGAAAAAAUGCCAAGGGGAGAUAGAAAUUGUGCAGCAAGC--------UCCAAGUGCAUGACAAACAGUGCAAUUGGGAGAGGCGGAGAGCAA .((((.((((((.((((((((((((.........((....)).....))))..))))))))..(--------(((...(((((........)))))....)))).)))))))))).. ( -45.24, z-score = -4.61, R) >droSec1.super_0 10427872 109 + 21120651 CGCUCAUCCGCCGUGUUGCACUUUCGAAAAAAUCCCAAGGGGAGAUAGAAAUUGUGCAGCAAGC--------UCCAAGUGCAUGACAAACAGUGCAAUUGGGAGAGGCGGAGAGCAA .((((.((((((.((((((((((((.......((((...))))....))))..))))))))..(--------(((...(((((........)))))....)))).)))))))))).. ( -49.50, z-score = -6.02, R) >droYak2.chr3R 13656487 107 + 28832112 CGCUCAUCCGCCGUGUUGCACUUUCGAAAAAAACGCAAGGGGAGCUAGAAAUUGUGCAGCA--C--------UCCAAGUGCAUGACAAACAGUGCAAUUGGAGGAGGUGGAGAGCAG .((((.((((((.((((((((((((........(....)........))))..))))))))--(--------(((((.(((((........))))).))))))..)))))))))).. ( -48.39, z-score = -5.70, R) >droEre2.scaffold_4770 10782374 106 - 17746568 CGCUCAUCCGCCGUGUUGCACUUUCG-AAAAAUCGCAAGGGGAGAUAGAAAUGGUGCAGCA--C--------UCCAAGUGCAUGACAAACAGUGCAAUUGGCAGAGGCGGAGAGCAG .((((.(((((((((((((((((((.-.....((....)).......)))..)))))))))--)--------.((((.(((((........))))).))))....)))))))))).. ( -49.22, z-score = -5.92, R) >droAna3.scaffold_13340 12840333 93 - 23697760 UACUCAUACGCCGUGUGGCGCUUUCGAAAAAAAUUAA---------AGAAAAUAUGCAGCA----------CUUCAAGUGCA--ACGGGGG---GUGGUGGGAGGAGAGAGCAGCGA ..(((((((.((.(((.(((.((((............---------.))))...))).(((----------(.....)))).--))).)).---)).)))))............... ( -21.32, z-score = 0.30, R) >dp4.chr2 20982722 102 + 30794189 ----------CGCUGUUGCACUUUCG-AAAAGCCAGAGACUUUUGUAGAAAAUAUGCGGCAGGCAACAGAGUAUCAAGUGCAUGACAAACA---UCAAUGAGAGGGGCGAAAAGAA- ----------..((((((((.((((.-(((((.......)))))...))))...))))))))((..(...((((...)))).(((......---)))......)..))........- ( -17.80, z-score = 1.32, R) >droPer1.super_0 10673511 102 - 11822988 ----------CGCUGUUGCACUUUCG-AAAAGCCAGAGACUUUUGUAGAAAAUAUGCGGCAGGCAACAGAGUAUCAAGUGCAUGACAAACA---UCAAAGAGAGGGGCGAAAAGAA- ----------..((((((((.((((.-(((((.......)))))...))))...))))))))((..(...((((...)))).(((......---)))......)..))........- ( -17.80, z-score = 1.16, R) >consensus CGCUCAUCCGCCGUGUUGCACUUUCG_AAAAAUCCCAAGGGGAGAUAGAAAUUGUGCAGCA_GC________UCCAAGUGCAUGACAAACAGUGCAAUUGGGAGAGGCGGAGAGCAA .............(((((((.((((......................))))...)))))))........................................................ ( -7.44 = -7.30 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:21 2011