
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,249,760 – 11,249,814 |
| Length | 54 |
| Max. P | 0.564762 |

| Location | 11,249,760 – 11,249,814 |
|---|---|
| Length | 54 |
| Sequences | 13 |
| Columns | 55 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
| Shannon entropy | 0.44110 |
| G+C content | 0.52399 |
| Mean single sequence MFE | -16.46 |
| Consensus MFE | -8.31 |
| Energy contribution | -8.34 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.564762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11249760 54 - 27905053 GAUCAGUACCUGCUGUCGUGGAACAACUUCCAUGGCAACAUGUGCCGCGGCUUC- .....((((.((.((((((((((....)))))))))).)).)))).........- ( -18.80, z-score = -2.19, R) >droSec1.super_0 10421855 54 + 21120651 GACCAGUACCUGCUGUCGUGGAACAACUUCCAUGGCAACAUGUGCCGCGGCUUC- ..((.((((.((.((((((((((....)))))))))).)).))))...))....- ( -19.30, z-score = -2.20, R) >droYak2.chr3R 13650312 54 + 28832112 GACCAGUACCUGUUGUCAUGGAACAACUUCCAUGGGAACAUGUGCCGCGGCUUC- ..((.((((.((((.((((((((....)))))))).)))).))))...))....- ( -19.70, z-score = -2.50, R) >droEre2.scaffold_4770 10776276 54 - 17746568 GACCAGUACCUGUUGUCGUGGAACAACUUCCAUGGGAACAUGUGCCGCGGCUUC- ..((.((((.((((.((((((((....)))))))).)))).))))...))....- ( -19.30, z-score = -2.04, R) >droAna3.scaffold_13340 12834446 54 - 23697760 GAUCAGUAUCUGCUGUCUUGGAACAACUUCCACGGGAACAUGUGCCGCGGCUUC- ...........(((((...((.(((..((((...))))..))).)))))))...- ( -11.20, z-score = 0.31, R) >dp4.chr2 20977158 54 + 30794189 GACCAGUAUCUGCUGUCGUGGAAUAACUUCCACGGCAACAUGUGCCGCGGCUUC- ..((.((((.((.((((((((((....)))))))))).)).))))...))....- ( -19.80, z-score = -2.78, R) >droPer1.super_0 10667832 54 - 11822988 GACCAGUAUCUGCUGUCGUGGAAUAACUUCCACGGCAACAUGUGCCGCGGCUUC- ..((.((((.((.((((((((((....)))))))))).)).))))...))....- ( -19.80, z-score = -2.78, R) >droWil1.scaffold_181089 2149644 54 - 12369635 GAUCAGUAUCUCCUGUCGUGGAACAAUUUCCACGGCAACAUGUGUCGCGGCUUU- .............((((((((((....)))))))))).................- ( -14.40, z-score = -1.36, R) >droVir3.scaffold_13047 5893728 54 + 19223366 GAUCAGUACCUGCUGUCGUGGAACAAUUUCCAUGGCAACAUGUGCCGCGGCUUU- .....((((.((.((((((((((....)))))))))).)).)))).........- ( -18.80, z-score = -2.23, R) >droMoj3.scaffold_6540 23586366 54 + 34148556 GAUCAGUAUCUGCUGUCGUGGAACAAUUUCCAUGGCAACAUGUGCCGCGGCUUC- .....((((.((.((((((((((....)))))))))).)).)))).........- ( -17.40, z-score = -1.90, R) >droGri2.scaffold_14906 4365073 54 - 14172833 GAUCAGUACCUAUUGUCGUGGAAUAACUUUCAUGGCAACAUGUGCCGUGGCUUU- .....((((...(((((((((((....)))))))))))...)))).........- ( -14.30, z-score = -1.44, R) >apiMel3.Group1 9947159 50 - 25854376 ----AAUUCUGCCUACGGUGGAACAAUUACCAGACGAAUCUGACCAACGUGUUC- ----..(((..((...))..))).......((((....))))............- ( -8.10, z-score = -0.02, R) >triCas2.ChLG10 1158525 54 + 8806720 GAGCAGUUUUCCCUCUGUUGGGACAAUUUCCACAAGAACAUGU-CCACGGGGAUG .........(((((......(((((..(((.....)))..)))-))..))))).. ( -13.10, z-score = -0.13, R) >consensus GAUCAGUACCUGCUGUCGUGGAACAACUUCCAUGGCAACAUGUGCCGCGGCUUC_ .....((((......((((((((....))))))))......)))).......... ( -8.31 = -8.34 + 0.03)

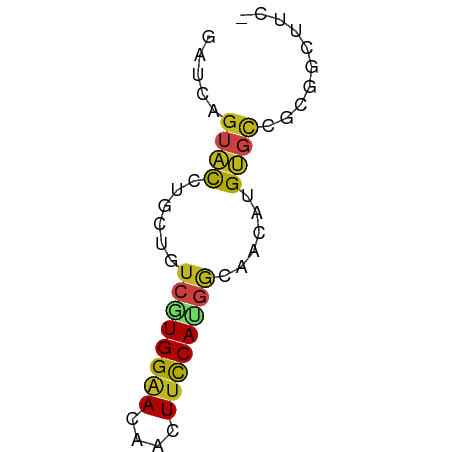
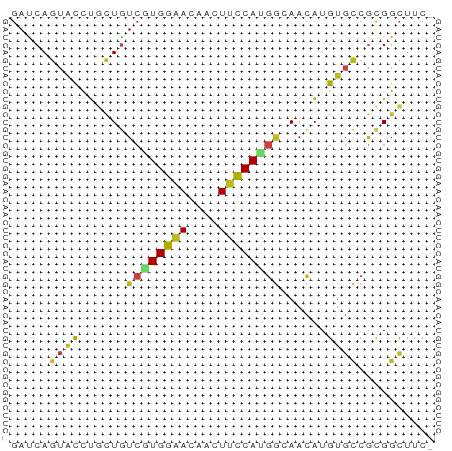
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:20 2011