| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,244,060 – 11,244,161 |
| Length | 101 |
| Max. P | 0.718558 |

| Location | 11,244,060 – 11,244,161 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.38 |
| Shannon entropy | 0.53533 |
| G+C content | 0.42289 |
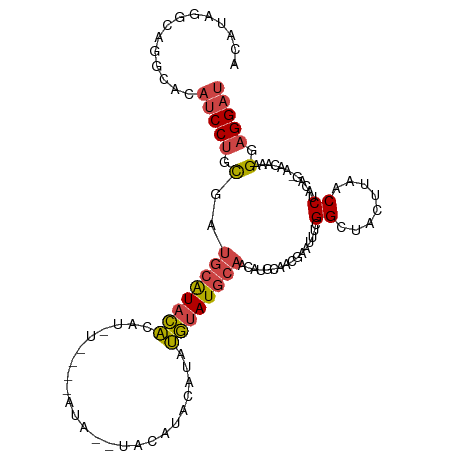
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -10.91 |
| Energy contribution | -11.24 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.718558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
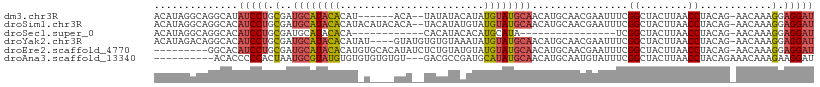
>dm3.chr3R 11244060 101 - 27905053 ACAUAGGCAGGCAUAUCCUGCGAUGCAUACACAU------ACA--UAUAUACAUAUGUAUGCAACAUGCAACGAAUUUCGGCUACUUAACCUACAG-AACAAAGGAGGAU ...((((((((.....)))))(.(((((...(((------(((--(((....)))))))))....))))).).........))).....(((.(..-......).))).. ( -23.10, z-score = -2.58, R) >droSim1.chr3R 17280282 107 + 27517382 ACAUAGGCAGGCACAUCCUGCGAUGCAUACACAUACAUACACA--UACAUAUGUAUGUAUGCAACAUGCAACGAAUUUCGGCUACUUAACCUACAG-AACAAAGGAGGAU ...((((((((.....)))))(.(((((...((((((((((..--......))))))))))....))))).).........))).....(((.(..-......).))).. ( -26.10, z-score = -3.04, R) >droSec1.super_0 10416293 81 + 21120651 ACAUAGGCAGGCACAUCCUGCGAUGCAUACACA------------CACAUACACAUGCAUA----------------UCGGCUACUUAACCUACAG-AACAAAGGAGGAU ...((((((((.....))))).((((((.....------------.........)))))).----------------....))).....(((.(..-......).))).. ( -13.74, z-score = -1.16, R) >droYak2.chr3R 13644234 105 + 28832112 ACAUAGACAGGCACAUCCUGCGAUGCAUACACAUAU----GUAUGUGUGUAAAUAUGUAUGCAACAUGCAACGAAUUUCGGCUACUUAACCUACAG-AACAAAGGAGGAU .........(((.(((..(((.(((((((((((((.----...))))))))....))))))))..)))...((.....)))))......(((.(..-......).))).. ( -20.00, z-score = 0.37, R) >droEre2.scaffold_4770 10770197 100 - 17746568 ---------GGCACAUCCUGCGAUGCAUACACAUGUGCACAUAUCUCUGUAUGUAUGUAUGCAACAUGCAACGAAUUUCGGCUACUUAACCUACAG-AACAAAGGAGGAU ---------.....(((((.((.((((((....))))))).....(((((((((((((.....))))))).((.....))...........)))))-).....).))))) ( -25.20, z-score = -1.37, R) >droAna3.scaffold_13340 12828426 97 - 23697760 ----------ACACCCCCACUAAUGCGUAUGUGUGUGUGUGU---GACGCCGAUGCAUAUGCAACAUGCAAUGUAUUUCGGCUACUUAACCUACAGAAACAAAGAAGGAU ----------................((.(.((((.((..((---(..((((((((((.(((.....))))))))..))))))))...)).)))).).)).......... ( -21.20, z-score = 0.12, R) >consensus ACAUAGGCAGGCACAUCCUGCGAUGCAUACACAU_U____AUA__UACAUACAUAUGUAUGCAACAUGCAACGAAUUUCGGCUACUUAACCUACAG_AACAAAGGAGGAU ..............(((((.(..((((((((........................))))))))................((........))............).))))) (-10.91 = -11.24 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:20 2011