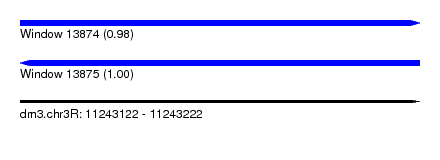
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,243,122 – 11,243,222 |
| Length | 100 |
| Max. P | 0.999377 |

| Location | 11,243,122 – 11,243,222 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.75 |
| Shannon entropy | 0.48614 |
| G+C content | 0.36176 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -11.24 |
| Energy contribution | -11.94 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980230 |
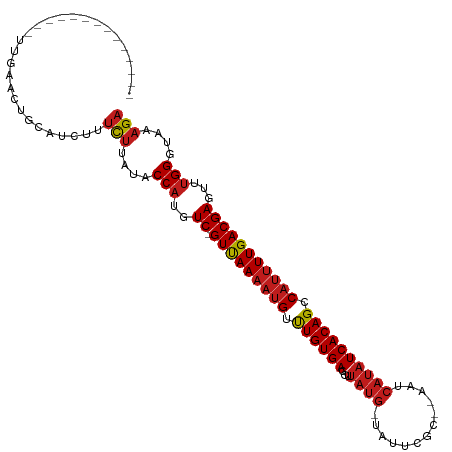
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11243122 100 + 27905053 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAUU--GUGAAUA-CAUAAGUUCACAAACAUUUUAAC-GACAUGGUAUAAGAAAGACGUGGUUCAGUCAG---------- ((((((((.....((((.((((((..(((((.....((--(((....-)))))..)))))..)))))).))-))...)))).))))..(((........)))..---------- ( -26.30, z-score = -2.76, R) >droSim1.chr3R 17279334 96 - 27517382 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAUU--GCGAAUA-CAUAAGUUCACAAACAUUUUAAC-GACAUGGUAUAAGAAAGACGCGGUUCAG-------------- ((((((((.....((((.((((((..(((((((((...--.......-))))...)))))..)))))).))-))...)))).))))..............-------------- ( -20.50, z-score = -1.64, R) >droSec1.super_0 10415372 96 - 21120651 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAUU--GCGAAUA-CAUAAGUUCACAAACAUUUUAAC-GACAUGGUAUAAGAAAGACGCGGUUCAG-------------- ((((((((.....((((.((((((..(((((((((...--.......-))))...)))))..)))))).))-))...)))).))))..............-------------- ( -20.50, z-score = -1.64, R) >droYak2.chr3R 13643301 96 - 28832112 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAUU--GCGAAUA-CAUAAGUUCACAAACAUUUUAAC-GACAUGGUAUAAGAAAGACGUGGGUCAA-------------- ((((((((.....((((.((((((..(((((((((...--.......-))))...)))))..)))))).))-))...)))).))))..(((....)))..-------------- ( -23.00, z-score = -2.12, R) >droEre2.scaffold_4770 10769265 96 + 17746568 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAUU--GCGAAUA-CAUAAGUUCACAAACAUUUUAAC-GACAUGGUAUAAGAAAGACGUUGUUCAA-------------- ((((((((.....((((.((((((..(((((((((...--.......-))))...)))))..)))))).))-))...)))).))))..............-------------- ( -20.50, z-score = -1.71, R) >droAna3.scaffold_13340 12827476 96 + 23697760 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAUU-GCGAAAUA-CAUAAGGUCACAAGCCUUUUAAC-GACAUGGUAUAAGAAAGAUGAACACAA--------------- ((((((((.....((((.((((.(((((((((((((((-....))).-)))...))))).)))))))).))-))...)))).)))).............--------------- ( -22.50, z-score = -2.68, R) >dp4.chr2 20969023 96 - 30794189 UCUGUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAUU-CAAGAGUA-CAUAAGGUCACAAACAUUUUAAC-GACAUGGUAUAAGAAAGAUGCUUUCCG--------------- ..((((((.....((((.((((((..((((((((((((-....))).-)))...))))))..)))))).))-))...)))))).(((((...)))))..--------------- ( -23.80, z-score = -2.64, R) >droPer1.super_0 10659928 96 + 11822988 UCUGUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAUU-CAAGAGUA-CAUAAGGUCACAAACAUUUUAAC-GACAUGGUAUAAGAAAGAUGCUUUCCG--------------- ..((((((.....((((.((((((..((((((((((((-....))).-)))...))))))..)))))).))-))...)))))).(((((...)))))..--------------- ( -23.80, z-score = -2.64, R) >droWil1.scaffold_181089 2143069 99 + 12369635 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAAUAGAGACACAACAUAAAGUCACAAACAUUUUAAC-GACAUGGUAUAAGAAAGUUGCAGUCAAU-------------- ((((((((.....((((.((((((..(((((((((.............)))...))))))..)))))).))-))...)))).))))..............-------------- ( -20.32, z-score = -2.06, R) >droVir3.scaffold_13047 5887499 105 - 19223366 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAU--AUAAAAUA-CAUAAGGUCACAGGCAUUUUAAC-GACAUGGUAUAAGAAAAUUGCAAUGCAAAAACGA-UAA---- ((((((((.....((((.((((((.(((((((...((--........-.))...))))))).)))))).))-))...)))).))))...((((...))))......-...---- ( -26.30, z-score = -4.00, R) >droMoj3.scaffold_6540 23579958 109 - 34148556 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAU--AGUGAAUA-CAUACGGUCACAGACAUUUUAAC-GACAUGGUAUAAGAAAAUUGCAAUUCGAAUGCAAAUGAAGA- ((((((((.....((((.((((((.(((((((...((--.((....)-)))...))))))).)))))).))-))...)))).))))...(((((.......))))).......- ( -27.20, z-score = -2.92, R) >droGri2.scaffold_14906 4357496 110 + 14172833 UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAU--AUAAAAUA-CAUAAGGUCACAAGCAUUUUAAC-GACAUGGUAUAAGAAAGAAAGAAAGAAACACACAAAGGAGAA ((((((((.....((((.((((((..((((((...((--........-.))...))))))..)))))).))-))...)))).))))............................ ( -19.20, z-score = -1.98, R) >triCas2.ChLG3 31519536 82 - 32080666 UCGUCAACCAAACUCGUCAAAAUGGCUGUGAUACCCGA--AUAAAUA-CUAGGUUUCACAACCAAAUUGACUGAUAGGGCACCGA----------------------------- (((....((....(((((((..(((.(((((.(((...--.......-...))).))))).)))..))))).))..))....)))----------------------------- ( -19.82, z-score = -2.10, R) >consensus UCUUUACCCAAACUCGUCAAAAUGGCUGUGAUAUGAUU__GAGAAUA_CAUAAGGUCACAAACAUUUUAAC_GACAUGGUAUAAGAAAGAUGCAGUUCAA______________ (((....(((...((((.((((((..(((((((((.............))))...)))))..)))))).)).))..)))....)))............................ (-11.24 = -11.94 + 0.70)

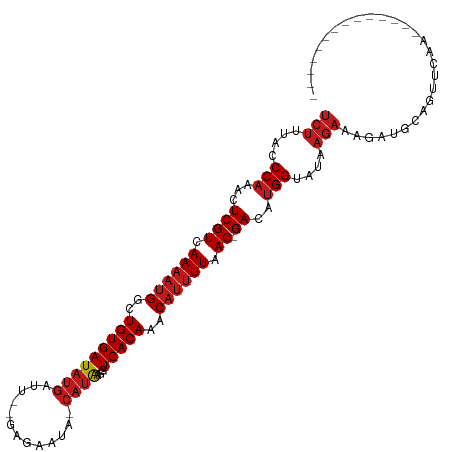
| Location | 11,243,122 – 11,243,222 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.75 |
| Shannon entropy | 0.48614 |
| G+C content | 0.36176 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.26 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
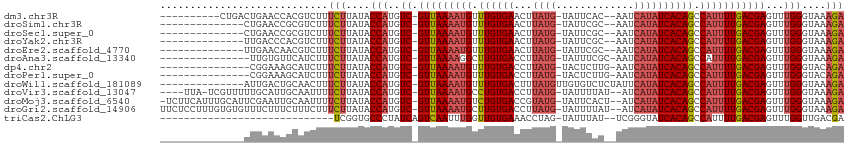
>dm3.chr3R 11243122 100 - 27905053 ----------CUGACUGAACCACGUCUUUCUUAUACCAUGUC-GUUAAAAUGUUUGUGAACUUAUG-UAUUCAC--AAUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA ----------..(((........)))..((((.((((...((-(((((((((.((((((...((((-.......--...)))))))))).))))))))))).....)))))))) ( -26.50, z-score = -2.83, R) >droSim1.chr3R 17279334 96 + 27517382 --------------CUGAACCGCGUCUUUCUUAUACCAUGUC-GUUAAAAUGUUUGUGAACUUAUG-UAUUCGC--AAUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA --------------..............((((.((((...((-(((((((((.((((((...((((-.......--...)))))))))).))))))))))).....)))))))) ( -24.90, z-score = -2.56, R) >droSec1.super_0 10415372 96 + 21120651 --------------CUGAACCGCGUCUUUCUUAUACCAUGUC-GUUAAAAUGUUUGUGAACUUAUG-UAUUCGC--AAUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA --------------..............((((.((((...((-(((((((((.((((((...((((-.......--...)))))))))).))))))))))).....)))))))) ( -24.90, z-score = -2.56, R) >droYak2.chr3R 13643301 96 + 28832112 --------------UUGACCCACGUCUUUCUUAUACCAUGUC-GUUAAAAUGUUUGUGAACUUAUG-UAUUCGC--AAUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA --------------..(((....)))..((((.((((...((-(((((((((.((((((...((((-.......--...)))))))))).))))))))))).....)))))))) ( -26.00, z-score = -3.19, R) >droEre2.scaffold_4770 10769265 96 - 17746568 --------------UUGAACAACGUCUUUCUUAUACCAUGUC-GUUAAAAUGUUUGUGAACUUAUG-UAUUCGC--AAUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA --------------..............((((.((((...((-(((((((((.((((((...((((-.......--...)))))))))).))))))))))).....)))))))) ( -24.90, z-score = -2.75, R) >droAna3.scaffold_13340 12827476 96 - 23697760 ---------------UUGUGUUCAUCUUUCUUAUACCAUGUC-GUUAAAAGGCUUGUGACCUUAUG-UAUUUCGC-AAUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA ---------------.............((((.((((...((-(((((((((((.((((...((((-.(((....-))))))))))))))).))))))))).....)))))))) ( -25.60, z-score = -2.91, R) >dp4.chr2 20969023 96 + 30794189 ---------------CGGAAAGCAUCUUUCUUAUACCAUGUC-GUUAAAAUGUUUGUGACCUUAUG-UACUCUUG-AAUCAUAUCACAGCCAUUUUGACGAGUUUGGGUACAGA ---------------.((((((...))))))....(((..((-(((((((((.((((((...((((-........-...)))))))))).)))))))))))...)))....... ( -25.50, z-score = -2.53, R) >droPer1.super_0 10659928 96 - 11822988 ---------------CGGAAAGCAUCUUUCUUAUACCAUGUC-GUUAAAAUGUUUGUGACCUUAUG-UACUCUUG-AAUCAUAUCACAGCCAUUUUGACGAGUUUGGGUACAGA ---------------.((((((...))))))....(((..((-(((((((((.((((((...((((-........-...)))))))))).)))))))))))...)))....... ( -25.50, z-score = -2.53, R) >droWil1.scaffold_181089 2143069 99 - 12369635 --------------AUUGACUGCAACUUUCUUAUACCAUGUC-GUUAAAAUGUUUGUGACUUUAUGUUGUGUCUCUAUUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA --------------..............((((.((((...((-(((((((((.((((((...((((..(((....))).)))))))))).))))))))))).....)))))))) ( -25.00, z-score = -2.53, R) >droVir3.scaffold_13047 5887499 105 + 19223366 ----UUA-UCGUUUUUGCAUUGCAAUUUUCUUAUACCAUGUC-GUUAAAAUGCCUGUGACCUUAUG-UAUUUUAU--AUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA ----...-......((((...))))...((((.((((...((-(((((((((.((((((...((((-........--..)))))))))).))))))))))).....)))))))) ( -28.10, z-score = -3.97, R) >droMoj3.scaffold_6540 23579958 109 + 34148556 -UCUUCAUUUGCAUUCGAAUUGCAAUUUUCUUAUACCAUGUC-GUUAAAAUGUCUGUGACCGUAUG-UAUUCACU--AUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA -.......(((((.......)))))...((((.((((...((-(((((((((.((((((...((((-........--..)))))))))).))))))))))).....)))))))) ( -31.10, z-score = -4.12, R) >droGri2.scaffold_14906 4357496 110 - 14172833 UUCUCCUUUGUGUGUUUCUUUCUUUCUUUCUUAUACCAUGUC-GUUAAAAUGCUUGUGACCUUAUG-UAUUUUAU--AUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA ............................((((.((((...((-(((((((((.((((((...((((-........--..)))))))))).))))))))))).....)))))))) ( -24.90, z-score = -3.59, R) >triCas2.ChLG3 31519536 82 + 32080666 -----------------------------UCGGUGCCCUAUCAGUCAAUUUGGUUGUGAAACCUAG-UAUUUAU--UCGGGUAUCACAGCCAUUUUGACGAGUUUGGUUGACGA -----------------------------(((..(((..((..(((((..(((((((((.(((..(-.......--)..))).)))))))))..)))))..))..)))...))) ( -32.90, z-score = -5.13, R) >consensus ______________UUGAACUGCAUCUUUCUUAUACCAUGUC_GUUAAAAUGUUUGUGACCUUAUG_UAUUCGC__AAUCAUAUCACAGCCAUUUUGACGAGUUUGGGUAAAGA ............................(((....(((..((.(((((((((.((((((...((((.............)))))))))).)))))))))))...)))....))) (-16.76 = -17.26 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:19 2011