
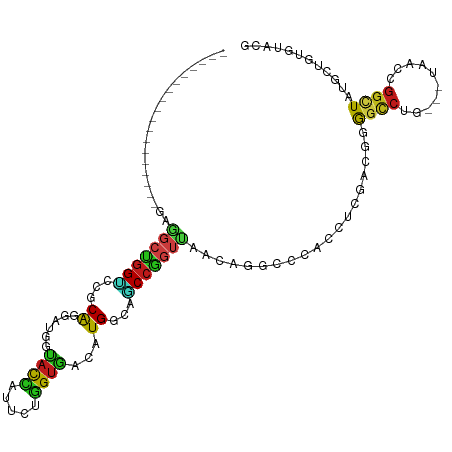
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,214,077 – 11,214,171 |
| Length | 94 |
| Max. P | 0.760750 |

| Location | 11,214,077 – 11,214,171 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 72.75 |
| Shannon entropy | 0.56548 |
| G+C content | 0.62084 |
| Mean single sequence MFE | -42.54 |
| Consensus MFE | -13.25 |
| Energy contribution | -12.88 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.94 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.760750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11214077 94 + 27905053 ------------------GAGGCCGGUCCGCAGGAUGGUACCAUUCUGGUGACAUGGCAGCCGGUUAACAGGCCCACCUCGACGGGGCCUG---UAACCGGCUAUGCUGUGUACG ------------------...((((((((.(((((((....)))))))).))).))))(((((((((.(((((((.........)))))))---)))))))))............ ( -49.50, z-score = -3.86, R) >droSec1.super_9 2621593 85 - 3197100 ---------------------GGCAGUUCGCGGGACGAUCUCACGG-GGAAGGAUGG--ACGGAGCCACGGGGACGACGAGAUCUGGCACU---UUGCCAAUUCCUGGAAGG--- ---------------------.....(((.(((((.((((((....-.......(((--......)))((....))..))))))((((...---..))))..))))))))..--- ( -28.20, z-score = -0.66, R) >droYak2.chr3R 13614234 94 - 28832112 ------------------GAGGCCGGUCCGCAGGAUGGUACCAUUCUGGUGACAUGGCAGCCGGUUAACAGGCCCACCUCAACGGGGCCUG---UAACCGGCUAUGCUGUGUACG ------------------...((((((((.(((((((....)))))))).))).))))(((((((((.(((((((.........)))))))---)))))))))............ ( -49.50, z-score = -4.02, R) >droEre2.scaffold_4770 10740045 94 + 17746568 ------------------GAGGCCGGUCCGCAGGACGGUACCAUUCUGGUGACAUGGCAGCCGGUUAACAGGCCCACCUCGACGGGGCCUG---UAACCGGCUAUGCUGUGUACG ------------------...((((.(((...)))))))(((.....))).((((((((((((((((.(((((((.........)))))))---))))))))..))))))))... ( -47.30, z-score = -3.14, R) >droAna3.scaffold_13340 12798524 94 + 23697760 ------------------GAGGCCGGCCCGCAGGACGGUACCAUUCUGGUGACAUGGCAGCCGGUUAACAGGCCCACCUCGACUGGGCCUG---UAACCGGCUAUGCUGUGUACG ------------------...((((.((....)).))))(((.....))).((((((((((((((((.((((((((.......))))))))---))))))))..))))))))... ( -48.60, z-score = -3.64, R) >dp4.chr2 20938053 94 - 30794189 ------------------GAGGCUGGCCCACAGGAUGGUACCAUUCUGGUGACAUGGCAGCCGGUCGCCAGGCCCACCGCGACUGGGCCUG---UUACCGGCUAUGCUGUGUACG ------------------(.((....))).(((((((....)))))))(((.(((((((((((((.((.(((((((.......))))))))---).))))))..)))))))))). ( -47.20, z-score = -2.58, R) >droPer1.super_0 10628789 94 + 11822988 ------------------GAGGCUGGCCCACAGGAUGGUACCAUUCUGGUGACAUGGCAGCCGGUCGCCAGGCCCACCGCGACUGAGCCUG---UUACCGGCUAUGCUGUGUACG ------------------..((((((((...((((((....))))))(((((((.(((...(((((((..((....))))))))).)))))---)))))))))).)))....... ( -42.50, z-score = -1.90, R) >droWil1.scaffold_181089 2107284 94 + 12369635 ------------------GAGGCUGGCCCUCAAGAUGGUACCAUUCUGGUGACAUGGCAGCCGGUUAACAGGCCCACCUCAACGGGGCCAG---UAACCGGCUAUGCUGUGUAUG ------------------((((.....)))).......((((.....))))((((((((((((((((...(((((.........)))))..---))))))))..))))))))... ( -41.30, z-score = -2.30, R) >droMoj3.scaffold_6540 23547754 94 - 34148556 ------------------GAGGCUGGUCCUCAGGAUGGCACCAUUCUGGUGACAUGGCAGCCGGUUAACCGACCCACGUCCACGGGGCCCG---UAACCGGCUACGCUGUGUACG ------------------((((.....))))((((((....)))))).(((.(((((((((((((((...(.(((.((....))))))...---)))))))))..))))))))). ( -41.00, z-score = -1.64, R) >droVir3.scaffold_13047 5857852 94 - 19223366 ------------------GAGGCUGGUCCUCAGGAUGGCACCAUUCUGGUGACAUGGCAGCCGGUUAACAGACCUACGUCCACGGGGCCUG---UAACCGGCUAUGCUGUGUAUG ------------------((((.....))))((((((....))))))....((((((((((((((((.(((.(((.((....))))).)))---))))))))..))))))))... ( -41.40, z-score = -2.46, R) >droGri2.scaffold_14906 4324590 94 + 14172833 ------------------GAGGCUGGCCCUCAGGAUGGUACCAUUCUGGUGACAUGGCAGCCGGUUAACAGACCCACGUCCACGGGGCCUG---UAACCGGCUAUGCUGUGUAUG ------------------((((.....))))((((((....))))))....((((((((((((((((.(((.(((.((....))))).)))---))))))))..))))))))... ( -44.60, z-score = -3.06, R) >anoGam1.chr2R 44661152 94 + 62725911 ------------------GAAGCUGGACCACAGGACGGUACGCUGCUCGUGACCUGGCAGCCGGU-GACGCGUCCACCUUCGUCCGGCCCG--GUGACCGGGUACGCCGUCUACG ------------------...(((((((....(((((....((((((........))))))((..-..)))))))......))))))).((--(((........)))))...... ( -37.00, z-score = 0.52, R) >apiMel3.GroupUn 354806058 106 - 399230636 UUAUAUUUGCAAUAGGUGGAGGCUGGACCGCAGGACGGCACUUUGCUAGUGACCUGGCAGCCUGUU-----GCCCUAAACGGUUCGGCC-G---UCACCGGUUACGCGGUGUAUG .((((((.((....((((..((((((((((.(((.(((((..(((((((....)))))))..))))-----).)))...))))))))))-.---.))))(....))))))))).. ( -47.40, z-score = -3.19, R) >triCas2.ChLG6 1307258 97 - 13544221 ------------------GAACCGGGGCCCCAGGAUGGUACUUUGCUUGUCACGUGGCACCCCAUUCUCACCCCCCACCACCCGGGGUCCACAAUAACCGGAUAUGCGGUUUAUG ------------------(((((((((....(((((((.....((((........))))..)))))))....)))).....((((............))))......)))))... ( -30.10, z-score = 0.56, R) >consensus __________________GAGGCUGGUCCGCAGGAUGGUACCAUUCUGGUGACAUGGCAGCCGGUUAACAGGCCCACCUCGACGGGGCCUG___UAACCGGCUAUGCUGUGUACG ....................(((((((...((......((((.....))))...))...)))))))...................((((..........))))............ (-13.25 = -12.88 + -0.37)


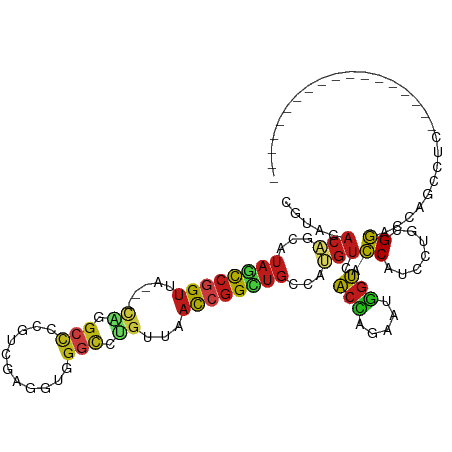
| Location | 11,214,077 – 11,214,171 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.75 |
| Shannon entropy | 0.56548 |
| G+C content | 0.62084 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -14.78 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.758528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11214077 94 - 27905053 CGUACACAGCAUAGCCGGUUA---CAGGCCCCGUCGAGGUGGGCCUGUUAACCGGCUGCCAUGUCACCAGAAUGGUACCAUCCUGCGGACCGGCCUC------------------ ............(((((((((---((((((((......).))))))).)))))))))(((..(((..(((.(((....))).)))..))).)))...------------------ ( -45.00, z-score = -3.50, R) >droSec1.super_9 2621593 85 + 3197100 ---CCUUCCAGGAAUUGGCAA---AGUGCCAGAUCUCGUCGUCCCCGUGGCUCCGU--CCAUCCUUCC-CCGUGAGAUCGUCCCGCGAACUGCC--------------------- ---..((((.(((..((((..---...))))(((((((.((.....((((......--))))......-.))))))))).))).).))).....--------------------- ( -21.90, z-score = -0.82, R) >droYak2.chr3R 13614234 94 + 28832112 CGUACACAGCAUAGCCGGUUA---CAGGCCCCGUUGAGGUGGGCCUGUUAACCGGCUGCCAUGUCACCAGAAUGGUACCAUCCUGCGGACCGGCCUC------------------ ............(((((((((---((((((((......).))))))).)))))))))(((..(((..(((.(((....))).)))..))).)))...------------------ ( -45.00, z-score = -3.49, R) >droEre2.scaffold_4770 10740045 94 - 17746568 CGUACACAGCAUAGCCGGUUA---CAGGCCCCGUCGAGGUGGGCCUGUUAACCGGCUGCCAUGUCACCAGAAUGGUACCGUCCUGCGGACCGGCCUC------------------ ............(((((((((---((((((((......).))))))).)))))))))(((..(((..(((.(((....))).)))..))).)))...------------------ ( -44.60, z-score = -3.02, R) >droAna3.scaffold_13340 12798524 94 - 23697760 CGUACACAGCAUAGCCGGUUA---CAGGCCCAGUCGAGGUGGGCCUGUUAACCGGCUGCCAUGUCACCAGAAUGGUACCGUCCUGCGGGCCGGCCUC------------------ ............(((((((((---((((((((.......)))))))).)))))))))(((..(((..(((.(((....))).)))..))).)))...------------------ ( -44.70, z-score = -2.72, R) >dp4.chr2 20938053 94 + 30794189 CGUACACAGCAUAGCCGGUAA---CAGGCCCAGUCGCGGUGGGCCUGGCGACCGGCUGCCAUGUCACCAGAAUGGUACCAUCCUGUGGGCCAGCCUC------------------ ....(((((...(((((((..---((((((((.......))))))))...)))))))(((((.((....)))))))......)))))((....))..------------------ ( -42.70, z-score = -1.74, R) >droPer1.super_0 10628789 94 - 11822988 CGUACACAGCAUAGCCGGUAA---CAGGCUCAGUCGCGGUGGGCCUGGCGACCGGCUGCCAUGUCACCAGAAUGGUACCAUCCUGUGGGCCAGCCUC------------------ ....(((((...(((((((..---((((((((.......))))))))...)))))))(((((.((....)))))))......)))))((....))..------------------ ( -39.70, z-score = -1.11, R) >droWil1.scaffold_181089 2107284 94 - 12369635 CAUACACAGCAUAGCCGGUUA---CUGGCCCCGUUGAGGUGGGCCUGUUAACCGGCUGCCAUGUCACCAGAAUGGUACCAUCUUGAGGGCCAGCCUC------------------ ........((.((((((((((---(.((((((......).))))).).))))))))))....(((..((..(((....)))..))..)))..))...------------------ ( -36.40, z-score = -1.33, R) >droMoj3.scaffold_6540 23547754 94 + 34148556 CGUACACAGCGUAGCCGGUUA---CGGGCCCCGUGGACGUGGGUCGGUUAACCGGCUGCCAUGUCACCAGAAUGGUGCCAUCCUGAGGACCAGCCUC------------------ ........(((((((((((((---(.(((((((....)).))))).).)))))))))))...(((..(((.(((....))).)))..)))..))...------------------ ( -43.80, z-score = -2.37, R) >droVir3.scaffold_13047 5857852 94 + 19223366 CAUACACAGCAUAGCCGGUUA---CAGGCCCCGUGGACGUAGGUCUGUUAACCGGCUGCCAUGUCACCAGAAUGGUGCCAUCCUGAGGACCAGCCUC------------------ ........((.((((((((((---((((((.((....))..)))))).))))))))))....(((..(((.(((....))).)))..)))..))...------------------ ( -38.00, z-score = -2.68, R) >droGri2.scaffold_14906 4324590 94 - 14172833 CAUACACAGCAUAGCCGGUUA---CAGGCCCCGUGGACGUGGGUCUGUUAACCGGCUGCCAUGUCACCAGAAUGGUACCAUCCUGAGGGCCAGCCUC------------------ ........((.((((((((((---(((((((((....)).))))))).))))))))))....(((..(((.(((....))).)))..)))..))...------------------ ( -42.70, z-score = -3.27, R) >anoGam1.chr2R 44661152 94 - 62725911 CGUAGACGGCGUACCCGGUCAC--CGGGCCGGACGAAGGUGGACGCGUC-ACCGGCUGCCAGGUCACGAGCAGCGUACCGUCCUGUGGUCCAGCUUC------------------ .((.(((.((...(((((...)--))))..(((((..(((((.....))-))).(((((..(....)..)))))....))))).)).)))..))...------------------ ( -37.40, z-score = 0.59, R) >apiMel3.GroupUn 354806058 106 + 399230636 CAUACACCGCGUAACCGGUGA---C-GGCCGAACCGUUUAGGGC-----AACAGGCUGCCAGGUCACUAGCAAAGUGCCGUCCUGCGGUCCAGCCUCCACCUAUUGCAAAUAUAA ..........((((..((((.---.-(((.(.(((((..(((((-----....((...)).((.((((.....)))))))))))))))).).)))..))))..))))........ ( -35.30, z-score = -1.20, R) >triCas2.ChLG6 1307258 97 + 13544221 CAUAAACCGCAUAUCCGGUUAUUGUGGACCCCGGGUGGUGGGGGGUGAGAAUGGGGUGCCACGUGACAAGCAAAGUACCAUCCUGGGGCCCCGGUUC------------------ (((...((((.(((((((............)))))))))))...))).((((.(((.(((..((.....))......((.....)))))))).))))------------------ ( -33.60, z-score = 1.05, R) >consensus CGUACACAGCAUAGCCGGUUA___CAGGCCCCGUCGAGGUGGGCCUGUUAACCGGCUGCCAUGUCACCAGAAUGGUACCAUCCUGCGGACCAGCCUC__________________ .....(((...((((((((.....((.(((...........))).))...))))))))...))).(((.....))).((.......))........................... (-14.78 = -14.96 + 0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:13 2011