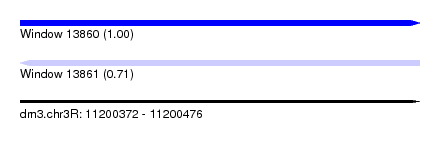
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,200,372 – 11,200,476 |
| Length | 104 |
| Max. P | 0.999433 |

| Location | 11,200,372 – 11,200,476 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.21 |
| Shannon entropy | 0.41565 |
| G+C content | 0.53519 |
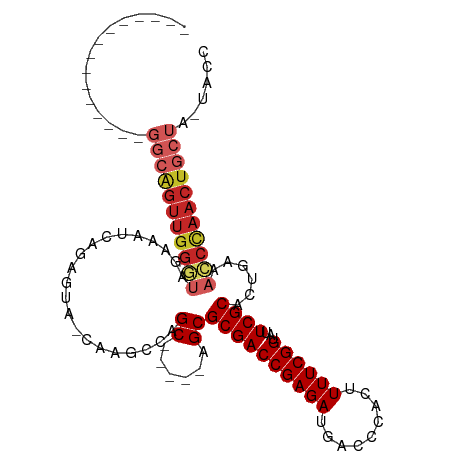
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -25.46 |
| Energy contribution | -26.66 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
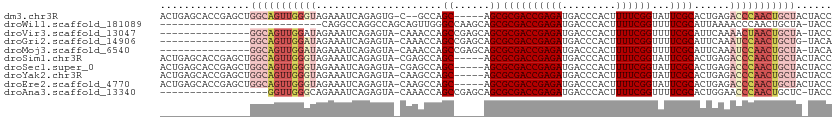
>dm3.chr3R 11200372 104 + 27905053 ACUGAGCACCGAGCUGGCAGUUGGGUAGAAAUCAGAGUG-C--GCCAGC-----AGCGCGACCGAGAUGACCCACUUUUCGGUAUUCGCACUGAGACCCAACUGCUACUACC ....(((.....)))(((((((((((.....((((..((-(--....))-----)(((..(((((((.........)))))))...))).)))).)))))))))))...... ( -41.80, z-score = -4.13, R) >droWil1.scaffold_181089 2094847 84 + 12369635 ---------------------------CAGGCCAGGCCAGCAGUUGGGCCAAGCAGCGCGACCGAGAUGACCCACUUUUCGGUUUUCGCAUUAAAACCCAACUGCUA-UACC ---------------------------..(((...)))((((((((((.......(((.((((((((.........))))))))..))).......)))))))))).-.... ( -33.04, z-score = -4.23, R) >droVir3.scaffold_13047 5845181 95 - 19223366 ---------------GGCAGUUGGAUAGAAAUCAGAGUA-CAAACCAGCCGAGCAGCGCGACCGAGAUGACCCACUUUUCGGUUUUCGCAUUCAAAACUAACUGCUA-UACC ---------------(((((((((...(....).((((.-.......((...)).(((.((((((((.........))))))))..)))))))....))))))))).-.... ( -26.20, z-score = -2.67, R) >droGri2.scaffold_14906 4310970 95 + 14172833 ---------------GGCAGUUGGAUAGAAAUCAGAGUA-CAAACCAGCCGAGCAGCGCGACCGAGAUGACCCACUUUUCGGUUUUCGCAUUCAAAUCCAACUGCUG-UACA ---------------(((((((((((.(....).((((.-.......((...)).(((.((((((((.........))))))))..)))))))..))))))))))).-.... ( -30.90, z-score = -3.46, R) >droMoj3.scaffold_6540 23535136 95 - 34148556 ---------------GGCAGUUGGAUAGAAAUCAGAGUA-CAAACCAGCCGAGCAGCGCGACCGAGAUGACCCACUUUUCGGUUUUCGCAUUCAAAUCCAACUGCUA-UACA ---------------(((((((((((.(....).((((.-.......((...)).(((.((((((((.........))))))))..)))))))..))))))))))).-.... ( -30.90, z-score = -4.18, R) >droSim1.chr3R 17245360 106 - 27517382 ACUGAGCACCGAGCUGGCAGUUGGGUAGAAAUCAGAGUA-CGAGCCAGC-----AGCGCGACCGAGAUGACCCACUUUUCGGUAUUCGCACUGAGACCCAACUGCUACUACC ....(((.....)))(((((((((((.....((((.((.-.......))-----.(((..(((((((.........)))))))...))).)))).)))))))))))...... ( -38.50, z-score = -3.50, R) >droSec1.super_0 10373239 106 - 21120651 ACUGAGCACCGAGCUGGCAGUUGGGUAGAAAUCAGAGUA-CGAGCCAGC-----AGCGCGACCGAGAUGACCCACUUUUCGGUAUUCGCACUGAGACCCAACUGCUACUACC ....(((.....)))(((((((((((.....((((.((.-.......))-----.(((..(((((((.........)))))))...))).)))).)))))))))))...... ( -38.50, z-score = -3.50, R) >droYak2.chr3R 13600195 106 - 28832112 ACUGAGCACCGAGCUGGCAGUUGGGUAGAAAUCAGAGUA-CAAGCCAGC-----AGCGCGACCGAGAUGACCCACUUUUCGGUAUUCGCACUGAGACCCAACUGCUACUACC ....(((.....)))(((((((((((.....((((.((.-.......))-----.(((..(((((((.........)))))))...))).)))).)))))))))))...... ( -38.50, z-score = -3.68, R) >droEre2.scaffold_4770 10726278 106 + 17746568 ACUGAGCACCGAGCUGGCAGUUGGGUAGAAAUCAGAGUA-CAAGCCAGC-----AGCGCGACCGAGAUGACCCACUUUUCGGUAUUCGCACUGAGACCCAACUGCUACUACC ....(((.....)))(((((((((((.....((((.((.-.......))-----.(((..(((((((.........)))))))...))).)))).)))))))))))...... ( -38.50, z-score = -3.68, R) >droAna3.scaffold_13340 12784635 92 + 23697760 ------------------GGUUGGGCAGAAAUCAGAGUA-CAAACCAGCCGAGCAGCGCGACCGAGAUGACCCACUUUUCGGUUUUCGCACUGGAACCCAACUGCUC-UACC ------------------((((((...............-....))))))((((((.((((((((((.........))))))...))))..(((...))).))))))-.... ( -29.91, z-score = -2.30, R) >consensus _______________GGCAGUUGGGUAGAAAUCAGAGUA_CAAGCCAGC_____AGCGCGACCGAGAUGACCCACUUUUCGGUAUUCGCACUGAAACCCAACUGCUA_UACC ...............(((((((((((.....................((......))((((((((((.........))))))...))))......)))))))))))...... (-25.46 = -26.66 + 1.20)
| Location | 11,200,372 – 11,200,476 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.21 |
| Shannon entropy | 0.41565 |
| G+C content | 0.53519 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -20.74 |
| Energy contribution | -21.92 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.710061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11200372 104 - 27905053 GGUAGUAGCAGUUGGGUCUCAGUGCGAAUACCGAAAAGUGGGUCAUCUCGGUCGCGCU-----GCUGGC--G-CACUCUGAUUUCUACCCAACUGCCAGCUCGGUGCUCAGU (((((...(((.((.(((.((((((((...((((.............)))))))))))-----)..)))--.-))..)))....)))))..((((..(((.....))))))) ( -36.42, z-score = -0.82, R) >droWil1.scaffold_181089 2094847 84 - 12369635 GGUA-UAGCAGUUGGGUUUUAAUGCGAAAACCGAAAAGUGGGUCAUCUCGGUCGCGCUGCUUGGCCCAACUGCUGGCCUGGCCUG--------------------------- (((.-(((((((((((((.....(((...(((((.............)))))..))).....))))))))))))).....)))..--------------------------- ( -35.42, z-score = -2.88, R) >droVir3.scaffold_13047 5845181 95 + 19223366 GGUA-UAGCAGUUAGUUUUGAAUGCGAAAACCGAAAAGUGGGUCAUCUCGGUCGCGCUGCUCGGCUGGUUUG-UACUCUGAUUUCUAUCCAACUGCC--------------- ....-..((((((......((.((((((..((((..(((((..(.....)..).))))..))))....))))-)).)).(((....))).)))))).--------------- ( -25.50, z-score = -0.67, R) >droGri2.scaffold_14906 4310970 95 - 14172833 UGUA-CAGCAGUUGGAUUUGAAUGCGAAAACCGAAAAGUGGGUCAUCUCGGUCGCGCUGCUCGGCUGGUUUG-UACUCUGAUUUCUAUCCAACUGCC--------------- ....-..((((((((((..((.((((((..((((..(((((..(.....)..).))))..))))....))))-)).))........)))))))))).--------------- ( -33.00, z-score = -2.83, R) >droMoj3.scaffold_6540 23535136 95 + 34148556 UGUA-UAGCAGUUGGAUUUGAAUGCGAAAACCGAAAAGUGGGUCAUCUCGGUCGCGCUGCUCGGCUGGUUUG-UACUCUGAUUUCUAUCCAACUGCC--------------- ....-..((((((((((..((.((((((..((((..(((((..(.....)..).))))..))))....))))-)).))........)))))))))).--------------- ( -33.00, z-score = -3.12, R) >droSim1.chr3R 17245360 106 + 27517382 GGUAGUAGCAGUUGGGUCUCAGUGCGAAUACCGAAAAGUGGGUCAUCUCGGUCGCGCU-----GCUGGCUCG-UACUCUGAUUUCUACCCAACUGCCAGCUCGGUGCUCAGU (((((...(((.((((((.((((((((...((((.............)))))))))))-----)..))))))-....)))....)))))..((((..(((.....))))))) ( -37.12, z-score = -1.24, R) >droSec1.super_0 10373239 106 + 21120651 GGUAGUAGCAGUUGGGUCUCAGUGCGAAUACCGAAAAGUGGGUCAUCUCGGUCGCGCU-----GCUGGCUCG-UACUCUGAUUUCUACCCAACUGCCAGCUCGGUGCUCAGU (((((...(((.((((((.((((((((...((((.............)))))))))))-----)..))))))-....)))....)))))..((((..(((.....))))))) ( -37.12, z-score = -1.24, R) >droYak2.chr3R 13600195 106 + 28832112 GGUAGUAGCAGUUGGGUCUCAGUGCGAAUACCGAAAAGUGGGUCAUCUCGGUCGCGCU-----GCUGGCUUG-UACUCUGAUUUCUACCCAACUGCCAGCUCGGUGCUCAGU (((((...(((.((((((.((((((((...((((.............)))))))))))-----)..))))))-....)))....)))))..((((..(((.....))))))) ( -34.42, z-score = -0.42, R) >droEre2.scaffold_4770 10726278 106 - 17746568 GGUAGUAGCAGUUGGGUCUCAGUGCGAAUACCGAAAAGUGGGUCAUCUCGGUCGCGCU-----GCUGGCUUG-UACUCUGAUUUCUACCCAACUGCCAGCUCGGUGCUCAGU (((((...(((.((((((.((((((((...((((.............)))))))))))-----)..))))))-....)))....)))))..((((..(((.....))))))) ( -34.42, z-score = -0.42, R) >droAna3.scaffold_13340 12784635 92 - 23697760 GGUA-GAGCAGUUGGGUUCCAGUGCGAAAACCGAAAAGUGGGUCAUCUCGGUCGCGCUGCUCGGCUGGUUUG-UACUCUGAUUUCUGCCCAACC------------------ ((((-(((((((((((...((((((((...((((.............))))))))))))))))))))((...-.))......))))))).....------------------ ( -31.82, z-score = -1.28, R) >consensus GGUA_UAGCAGUUGGGUCUCAGUGCGAAAACCGAAAAGUGGGUCAUCUCGGUCGCGCU_____GCUGGCUUG_UACUCUGAUUUCUACCCAACUGCC_______________ .......((((((((((......((((...((((.............))))))))((......)).....................))))))))))................ (-20.74 = -21.92 + 1.18)

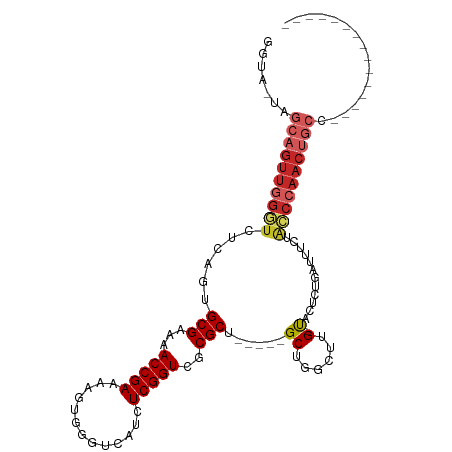
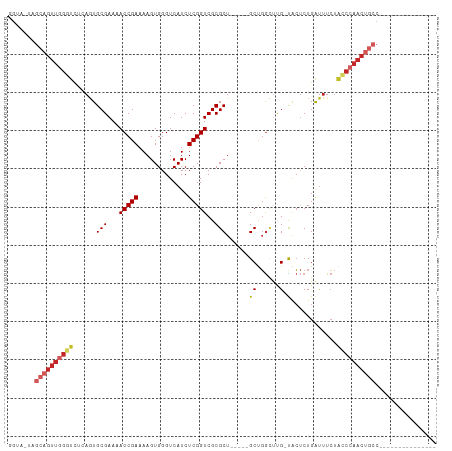
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:06 2011