| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,184,035 – 11,184,129 |
| Length | 94 |
| Max. P | 0.996121 |

| Location | 11,184,035 – 11,184,129 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 62.36 |
| Shannon entropy | 0.74368 |
| G+C content | 0.42328 |
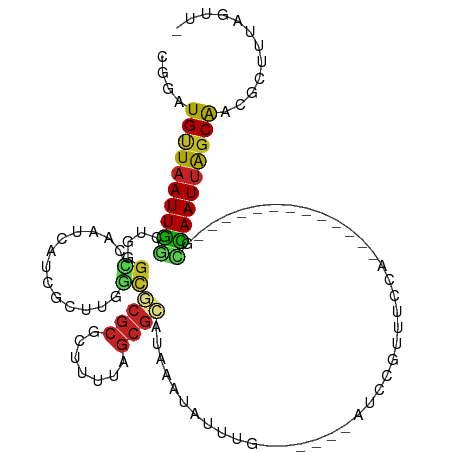
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -10.26 |
| Energy contribution | -9.86 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.69 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
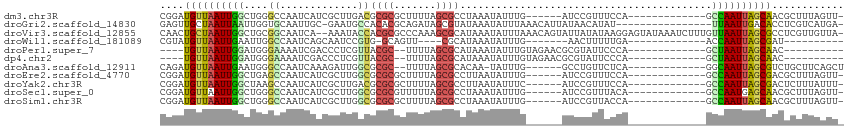
>dm3.chr3R 11184035 94 + 27905053 CGGAUGUUAAUUGGCUGGGCCAAUCAUCGCUUGACGCGCGCUUUUAGCGCCUAAAUAUUUG------AUCCGUUUCCA-------------GCCAAUUAGCAACGCUUUAGUU- ((..((((((((((((((((..((((..((.....))((((.....)))).........))------))..))..)))-------------))))))))))).))........- ( -33.00, z-score = -3.09, R) >droGri2.scaffold_14830 6094705 96 - 6267026 GAGUUGCUAAUUAAUUGGUGCAAUUGC-GAAUGCCACACGCAGAUAGCGUAUAAAUAUUUAAACAUUAUAACAUAU----------------UUAAUUGACACCUCGUCAUGA- (((.((.(((((((.((((((....))-....)))).((((.....))))..........................----------------))))))).)).))).......- ( -15.90, z-score = 0.14, R) >droVir3.scaffold_12855 8375665 111 - 10161210 CAACUGCUAAUUGGCUGCGGCAAUCA--AAAUACCACGCGCCCAAAGCGCAUAAAUAUUUAAACAGUAUUAUAUAAGGAGUAUAAAUCUUUGUUAAUUAGCGCCUCGUUGUUA- (((((((((((((((((.((......--.....))..((((.....)))).............)))).....(((((((.......))))))))))))))))....))))...- ( -23.10, z-score = -0.41, R) >droWil1.scaffold_181089 4013079 79 + 12369635 CGUAUGUUAAUUGAAUUGGCCAAUCAGCAAUCCGUG-GCAGUU----CGCAUAAAUAUUUG-------AACUUUUUGA-------------ACCAAUUAGCGAU---------- ....(((((((((.....((((...(....)...))-))((((----((.((....)).))-------))))......-------------..)))))))))..---------- ( -16.60, z-score = -0.96, R) >droPer1.super_7 4215384 85 - 4445127 ----UGUUAAUUGGAUGGGAAAAUCGACCCUCGUUACGC--UUUUAGCGCAUAAAUAUUUGUAGAACGCGUAUUCCCA-------------GCUAAUUAGCAAC---------- ----((((((((((.((((((...((.....)).(((((--.(((.(((.((....)).))).))).)))))))))))-------------.))))))))))..---------- ( -26.50, z-score = -3.73, R) >dp4.chr2 4125150 85 + 30794189 ----UGUUAAUUGGAUGGGAAAAUCGACCCUCGUUACGC--UUUUAGCGCAUAAAUAUUUGUAGAACGCGUAUUCCCA-------------GCUAAUUAGCAAC---------- ----((((((((((.((((((...((.....)).(((((--.(((.(((.((....)).))).))).)))))))))))-------------.))))))))))..---------- ( -26.50, z-score = -3.73, R) >droAna3.scaffold_12911 1970789 92 - 5364042 CAGAUGUUAAUUGAAUGGGCCAAUCAAAGAUUGGCGCGC--UUUUAGCGCACAA-UAUUUG------GCCUGUUCUCA-------------GGCAAUUAGCGUCUGCUUCAGCU (((((((((((((((((.(((((((...)))))))((((--.....))))....-))))).------(((((....))-------------)))))))))))))))........ ( -38.70, z-score = -4.71, R) >droEre2.scaffold_4770 10709521 94 + 17746568 CGGAUGUUAAUUGGCUGAGCCAAUCAUCGCUUGGCGCGCGCUUUUAGCGCCUUAAUAUUUG------AUCCGUUUCCA-------------GCCAAUUAGCGACGCUUUAGUU- ((..(((((((((((((.(((((.......)))))((((.......))))...........------.........))-------------))))))))))).))........- ( -32.10, z-score = -2.37, R) >droYak2.chr3R 13583533 94 - 28832112 CGGAUGUUAAUUGGCUAAGCCAAUCAUCGCUUGACGCGCGCUUUUAGCGCCUUAAUAUUUC------AUCCGUUUCCA-------------GCCAAUUAGCGACUCUUUAUUU- .((.((((((((((((((((........))))...((((.......))))...........------..........)-------------))))))))))).))........- ( -25.10, z-score = -2.26, R) >droSec1.super_0 10356773 94 - 21120651 CGGAUGUUAAUUGGCUGGGCCAAUCAUCGCUUGGCGCGCGUUUUUAGCGCCUAAAUAUUUG------AUCCGUUUACA-------------GCCAAUGAGCAACGCUUUAGUU- ((..((((.((((((((.(((((.......)))))((((.......))))...........------.........))-------------)))))).)))).))........- ( -29.10, z-score = -1.25, R) >droSim1.chr3R 17229176 94 - 27517382 CGGAUGUUAAUUGGCUGGGCCAAUCAUCGCUUGGCGCGCGCUUUUAGCGCCUAAAUAUUUG------AUCCGUUACCA-------------GCCAAUUAGCAACGCUUUAGUU- ((..(((((((((((((((((((.......)))))((((.......))))...........------........)))-------------))))))))))).))........- ( -36.30, z-score = -3.44, R) >consensus CGGAUGUUAAUUGGCUGGGCCAAUCAUCGCUUGGCGCGCGCUUUUAGCGCAUAAAUAUUUG______AUCCGUUUCCA_____________GCCAAUUAGCAACGCUUUAGUU_ ....((((((((((....((.............))((((.......))))..........................................))))))))))............ (-10.26 = -9.86 + -0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:05 2011