| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,122,307 – 11,122,435 |
| Length | 128 |
| Max. P | 0.900410 |

| Location | 11,122,307 – 11,122,435 |
|---|---|
| Length | 128 |
| Sequences | 15 |
| Columns | 138 |
| Reading direction | reverse |
| Mean pairwise identity | 90.84 |
| Shannon entropy | 0.21072 |
| G+C content | 0.51938 |
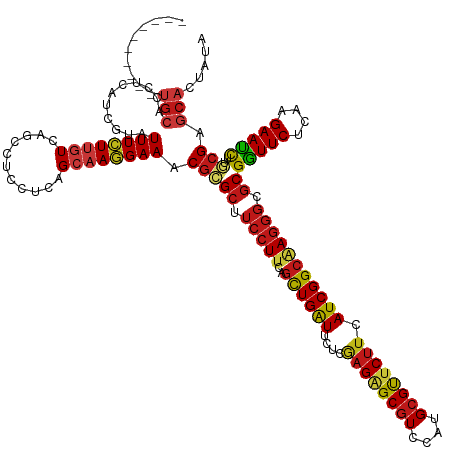
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -32.74 |
| Energy contribution | -32.99 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
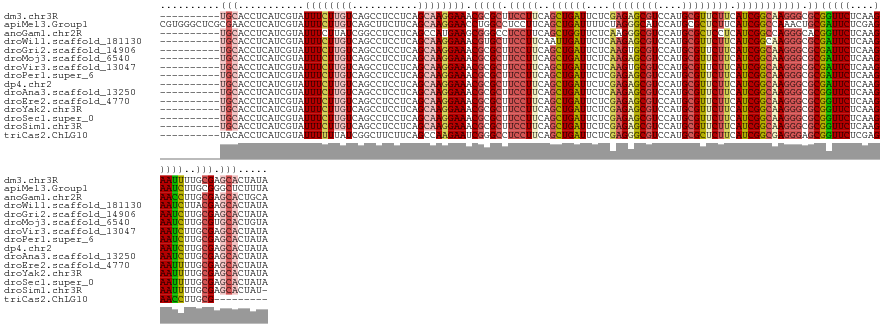
>dm3.chr3R 11122307 128 - 27905053 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAUA ----------.........(((((.((((((..((((((((.....))))...((((..((((..((((((....((((((((....)))))))).))))))))))))))))))..))))))...)))))........ ( -44.90, z-score = -2.95, R) >apiMel3.Group1 10372741 138 + 25854376 CGUGGGCUCGCGAACCUCAUCGUAUUUCUUGUCAGCUUCUUCAGCAAGGAACCUGGCCUCCUUCAGCUGAUUUUCUAGGGCAUCCAUGCGCUCUUCAUCGGCCAAACUGCGAUUCUCGAGAAUCUUGCGGGCUCUUUA ...((((((((((..(((((((((......(((((.(((((.....))))).)))))........((((((.....(((((........)))))..)))))).....))))))....)))....)))))))))).... ( -42.50, z-score = -1.16, R) >anoGam1.chr2R 10564783 128 - 62725911 ----------UGCACCUCAUCGUAUUUCUUAUCGGCCUCCUCAGCCAUGAAGCGGGCCUCCUUCAGCUGGUUCUCAAGGGCGUCCAUGCGCUCCUCAUCGGCCAGGGCACGGUUCUCAAGAACCUUGCGAGCACUGCA ----------.(((.(((...............(((((.((((....)).)).)))))........((((((....(((((((....)))).)))....)))))).(((.(((((....))))).))))))...))). ( -43.00, z-score = -0.97, R) >droWil1.scaffold_181130 3658985 128 - 16660200 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGUGCUUCCUUCAAUUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUACGAGCACUAUA ----------...........((.((((((((...........))))))))))((((((.........((((((..(((((((((.(((...........))).))))))...)))..))))))....)))))).... ( -33.82, z-score = -1.50, R) >droGri2.scaffold_14906 884823 128 - 14172833 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGUGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGAGCACUAUA ----------.........(((((.((((((.........(((((((((((......))))))..)))))........(((((((.(((...........))).))))))).....))))))...)))))........ ( -36.70, z-score = -1.29, R) >droMoj3.scaffold_6540 18587641 128 + 34148556 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGUGCACUGUA ----------(((((...........(((((((.......(((((((((((......))))))..))))).....((((((((....))))))))....)))))))(((.(((((....))))).))))))))..... ( -42.40, z-score = -2.60, R) >droVir3.scaffold_13047 11383726 128 + 19223366 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGUGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGAGCACUAUA ----------.........(((((.((((((.........(((((((((((......))))))..)))))........(((((((.(((...........))).))))))).....))))))...)))))........ ( -36.70, z-score = -1.29, R) >droPer1.super_6 3375591 128 - 6141320 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGAGCACUAUA ----------.........(((((.((((((...((.......))..((((.(((((..((((..((((((....((((((((....)))))))).))))))))))))))).))))))))))...)))))........ ( -41.00, z-score = -1.85, R) >dp4.chr2 28049178 128 - 30794189 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGAGCACUAUA ----------.........(((((.((((((...((.......))..((((.(((((..((((..((((((....((((((((....)))))))).))))))))))))))).))))))))))...)))))........ ( -41.00, z-score = -1.85, R) >droAna3.scaffold_13250 3403786 128 + 3535662 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUCUUGCGAGCACUAUA ----------.........(((((.((((((..((((((((.....))))...((((..((((..((((((....((((((((....)))))))).))))))))))))))))))..))))))...)))))........ ( -44.50, z-score = -3.32, R) >droEre2.scaffold_4770 10645982 128 - 17746568 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAUA ----------.........(((((.((((((..((((((((.....))))...((((..((((..((((((....((((((((....)))))))).))))))))))))))))))..))))))...)))))........ ( -44.90, z-score = -2.95, R) >droYak2.chr3R 13521027 128 + 28832112 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAUA ----------.........(((((.((((((..((((((((.....))))...((((..((((..((((((....((((((((....)))))))).))))))))))))))))))..))))))...)))))........ ( -44.90, z-score = -2.95, R) >droSec1.super_0 10291021 128 + 21120651 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAUA ----------.........(((((.((((((..((((((((.....))))...((((..((((..((((((....((((((((....)))))))).))))))))))))))))))..))))))...)))))........ ( -44.90, z-score = -2.95, R) >droSim1.chr3R 17173441 127 + 27517382 ----------UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAU- ----------.........(((((.((((((..((((((((.....))))...((((..((((..((((((....((((((((....)))))))).))))))))))))))))))..))))))...))))).......- ( -44.90, z-score = -2.83, R) >triCas2.ChLG10 353869 119 + 8806720 ----------UACACCUCAUCGUAUUUUUUAUCGGCUUCUUCAGCCAAGAAUCGGGCCUCCUUCAGCUGAUUCUCGAGGGCGUCCAUGCGCUCUUCAUCGGCGAGGGAGCGGUUCUCGAGAACCUUGCG--------- ----------.....(((...............((((.....)))).(((((((...(((((((.((((((....((((((((....)))))))).)))))))))))))))))))).))).........--------- ( -42.90, z-score = -2.22, R) >consensus __________UGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUCUUGCGAGCACUAUA ..........(((...........((((((((...........)))))))).(((((.(((((..((((((....((((((((....)))))))).))))))))))).))(((((....)))))..))).)))..... (-32.74 = -32.99 + 0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:02 2011