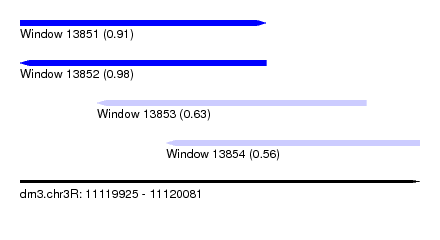
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,119,925 – 11,120,081 |
| Length | 156 |
| Max. P | 0.975036 |

| Location | 11,119,925 – 11,120,021 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 62.82 |
| Shannon entropy | 0.65214 |
| G+C content | 0.48094 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -13.66 |
| Energy contribution | -12.58 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
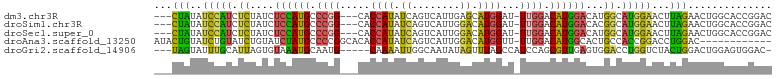
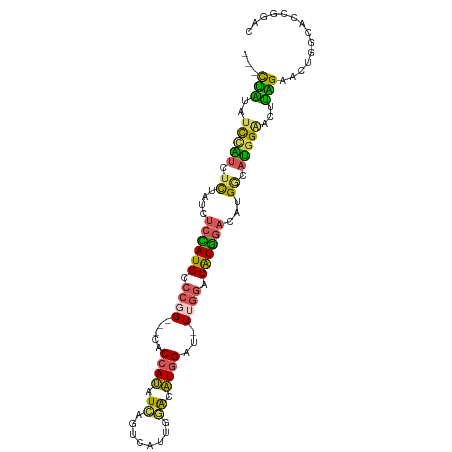
>dm3.chr3R 11119925 96 + 27905053 ---CUAUAUCCAUCUCUAUCUCCAUGCCCGG---CACCAUAUCAGUCAUUGAGCAUGGAU-UUGGACAUGGACAUGGCAUGGAACUUAGAACUGGCACCGGAC ---(((..(((((..((((.((((((.((((---..((((.((((...))))..))))..-)))).)))))).)))).)))))...)))..(((....))).. ( -29.00, z-score = -1.53, R) >droSim1.chr3R 17171043 96 - 27517382 ---CUAUAUCCAUCUCUAUCUCCAUGCCCGG---CACCAUAUCAGUCAUUGGACAUGGAU-UUGGACAUGGACACGGCAUGGAACUUAGAACUGGCACCGGAC ---......(((..((((..((((((((.((---(.........)))..((..((((...-.....))))..)).))))))))...))))..)))........ ( -28.70, z-score = -1.80, R) >droSec1.super_0 10288624 96 - 21120651 ---CUAUAUCCAUCUCUAUCUCCAUGCCCGG---CACCAUAUCAGUCAUUGGACAUGGAU-UUGGACAUGGACAUGGCAUGGAACUUAGAACUGGCACCGGAC ---(((..(((((..((((.((((((.((((---..((((.((........)).))))..-)))).)))))).)))).)))))...)))..(((....))).. ( -28.70, z-score = -1.46, R) >droAna3.scaffold_13250 3401204 90 - 3535662 AUACUGUAUCUGUAUCUGUAUCUAUGCCCCGGCACACCAUAUCAGUCAUUGGACAUGGUU-UUGGACAUGGCACUGCCACCGGACCUGGAC------------ ((((.......)))).....((((.(..((((....(((.(((((((....))).)))).-.)))...((((...))))))))..))))).------------ ( -20.20, z-score = 1.35, R) >droGri2.scaffold_14906 882221 94 + 14172833 ---UAGUAUUUGCAUUAGUGUAAAUGCAAUG-----CAAAAUUGGCAAUAUAGUUUAGCCAUCCAGCGUUGAGUGGACCUGGUCUACUGGACUGGAGUGGAC- ---..(((((((((....)))))))))..((-----(.......)))...........(((((((..(((.(((((((...))))))).))))))).)))..- ( -29.20, z-score = -2.18, R) >consensus ___CUAUAUCCAUCUCUAUCUCCAUGCCCGG___CACCAUAUCAGUCAUUGGACAUGGAU_UUGGACAUGGACAUGGCAUGGAACUUAGAACUGGCACCGGAC ...(((..(((((.((....((((((.((((.....((((.((........)).))))...)))).))))))...)).)))))...))).............. (-13.66 = -12.58 + -1.08)
| Location | 11,119,925 – 11,120,021 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 62.82 |
| Shannon entropy | 0.65214 |
| G+C content | 0.48094 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -14.76 |
| Energy contribution | -14.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.975036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11119925 96 - 27905053 GUCCGGUGCCAGUUCUAAGUUCCAUGCCAUGUCCAUGUCCAA-AUCCAUGCUCAAUGACUGAUAUGGUG---CCGGGCAUGGAGAUAGAGAUGGAUAUAG--- .....((((((.(((((..(((((((((..(((((((((...-...(((.....)))...))))))).)---)..))))))))).))))).))).)))..--- ( -32.30, z-score = -1.77, R) >droSim1.chr3R 17171043 96 + 27517382 GUCCGGUGCCAGUUCUAAGUUCCAUGCCGUGUCCAUGUCCAA-AUCCAUGUCCAAUGACUGAUAUGGUG---CCGGGCAUGGAGAUAGAGAUGGAUAUAG--- .....((((((.(((((..(((((((((..(((((((((...-...(((.....)))...))))))).)---)..))))))))).))))).))).)))..--- ( -31.90, z-score = -1.91, R) >droSec1.super_0 10288624 96 + 21120651 GUCCGGUGCCAGUUCUAAGUUCCAUGCCAUGUCCAUGUCCAA-AUCCAUGUCCAAUGACUGAUAUGGUG---CCGGGCAUGGAGAUAGAGAUGGAUAUAG--- .....((((((.(((((..(((((((((..(((((((((...-...(((.....)))...))))))).)---)..))))))))).))))).))).)))..--- ( -32.30, z-score = -1.98, R) >droAna3.scaffold_13250 3401204 90 + 3535662 ------------GUCCAGGUCCGGUGGCAGUGCCAUGUCCAA-AACCAUGUCCAAUGACUGAUAUGGUGUGCCGGGGCAUAGAUACAGAUACAGAUACAGUAU ------------((((.((..(.(((((...)))))......-.((((((((........)))))))))..)).))))..........((((.......)))) ( -25.30, z-score = 0.24, R) >droGri2.scaffold_14906 882221 94 - 14172833 -GUCCACUCCAGUCCAGUAGACCAGGUCCACUCAACGCUGGAUGGCUAAACUAUAUUGCCAAUUUUG-----CAUUGCAUUUACACUAAUGCAAAUACUA--- -......((((((..(((.(((...))).)))....))))))((((...........))))......-----..(((((((......)))))))......--- ( -21.60, z-score = -1.85, R) >consensus GUCCGGUGCCAGUUCUAAGUUCCAUGCCAUGUCCAUGUCCAA_AUCCAUGUCCAAUGACUGAUAUGGUG___CCGGGCAUGGAGAUAGAGAUGGAUAUAG___ ...............................(((((((((.....(((((((........))))))).......))))))))).................... (-14.76 = -14.76 + -0.00)
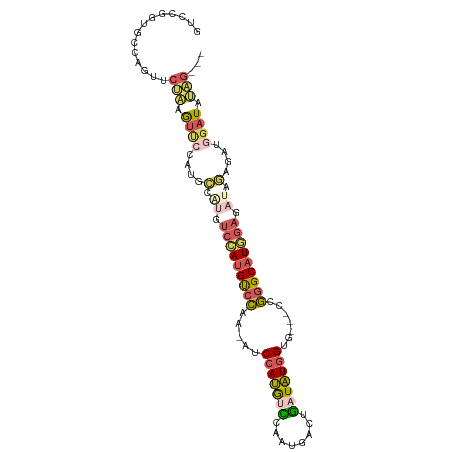
| Location | 11,119,955 – 11,120,060 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 70.01 |
| Shannon entropy | 0.54560 |
| G+C content | 0.47135 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -11.78 |
| Energy contribution | -13.02 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11119955 105 - 27905053 UUUGAAUCG-UAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUGCCAUGUCCAUGUCCAA-AUCCAUGCUCAAUGACUGAUAUGG (((((..((-....))..)))))((((..((((((.((((....((.((.........)).)).)))).))))))))))...-..(((((.(((.....)))))))) ( -30.20, z-score = -1.66, R) >droSim1.chr3R 17171073 105 + 27517382 UUUGAAUCG-AAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUGCCGUGUCCAUGUCCAA-AUCCAUGUCCAAUGACUGAUAUGG (((((..((-....))..)))))((((..((((((.((((....((.((.........)).)).)))).))))))))))...-..(((((((........))))))) ( -31.40, z-score = -1.96, R) >droSec1.super_0 10288654 105 + 21120651 AUUGAAUCG-AAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUGCCAUGUCCAUGUCCAA-AUCCAUGUCCAAUGACUGAUAUGG .((((..((-....))..)))).((((..((((((.((((....((.((.........)).)).)))).))))))))))...-..(((((((........))))))) ( -30.80, z-score = -1.80, R) >droAna3.scaffold_13250 3401240 94 + 3535662 UUUAAAUCGAAAAAAGGCAGAAAAGCAAUUGGACAAGGCAGUCC------------AGGUCCGGUGGCAGUGCCAUGUCCAA-AACCAUGUCCAAUGACUGAUAUGG ................((......))..(((((((.((((.(((------------(.......))).).)))).)))))))-..(((((((........))))))) ( -28.20, z-score = -2.29, R) >droGri2.scaffold_14906 882251 98 - 14172833 CUUUGACCA-AAUGCGGC-----AUCGAUUGGACAAGGCAGUCCACU-CCAGUCCAGUAGACCAGGUCCACUCAACGCUGGAUGGCUAAACUAUAUUGCCAAUUU-- ..(((.(((-(...((..-----..)).)))).)))(((((((((.(-(((((..(((.(((...))).)))....))))))))).........)))))).....-- ( -24.90, z-score = -0.59, R) >consensus UUUGAAUCG_AAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUGCCAUGUCCAUGUCCAA_AUCCAUGUCCAAUGACUGAUAUGG .......................((((..((((((.((((........................)))).))))))))))......(((((((........))))))) (-11.78 = -13.02 + 1.24)

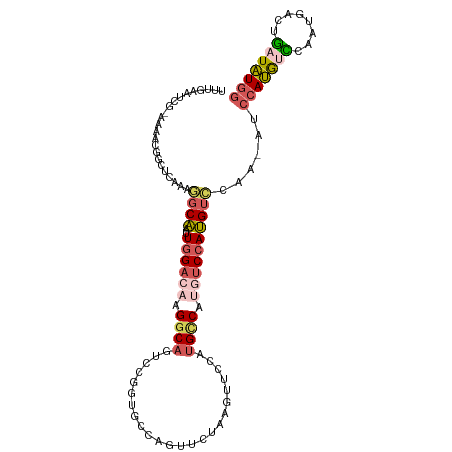
| Location | 11,119,982 – 11,120,081 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.55 |
| Shannon entropy | 0.38008 |
| G+C content | 0.45382 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -13.85 |
| Energy contribution | -15.72 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11119982 99 - 27905053 AAAAUUAGAGGCGAAUCAAACUUUGAAUCG-UAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUGCCAUGUCCAUGUC .........((((.......((((((..((-....))..)))))).....((((((.((((....((.((.........)).)).)))).)))))))))) ( -27.80, z-score = -2.03, R) >droSim1.chr3R 17171100 99 + 27517382 AAAAUUAGAGGCGAAUCAAACUUUGAAUCG-AAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUGCCGUGUCCAUGUC .........((((.......((((((..((-....))..)))))).....((((((.((((....((.((.........)).)).)))).)))))))))) ( -27.00, z-score = -1.47, R) >droSec1.super_0 10288681 99 + 21120651 AAAAUUAGAGGCGAAUCAAACAUUGAAUCG-AAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUGCCAUGUCCAUGUC .........(((((((..(((.((((..((-....))..)))).((((.((((((......)))))))))).)))....))))...)))........... ( -25.00, z-score = -1.15, R) >droAna3.scaffold_13250 3401267 77 + 3535662 -----------AAAGUAUAAGUUUAAAUCGAAAAAAGGCAGAAAAGCAAUUGGACAAGGCAGUCCAGGUCCGGUGGC-AGUGCCAUGUC----------- -----------.........................((((.....((.(((((((..((....))..))))))).))-..)))).....----------- ( -17.70, z-score = -1.96, R) >consensus AAAAUUAGAGGCGAAUCAAACUUUGAAUCG_AAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUGCCAUGUCCAUGUC .........((((........(((((.((.......)).)))))((((.((((((......))))))))))..............))))........... (-13.85 = -15.72 + 1.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:17:00 2011