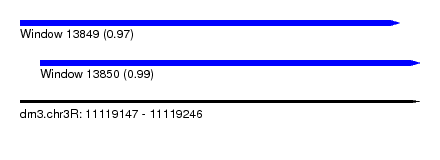
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,119,147 – 11,119,246 |
| Length | 99 |
| Max. P | 0.991057 |

| Location | 11,119,147 – 11,119,241 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Shannon entropy | 0.43211 |
| G+C content | 0.39900 |
| Mean single sequence MFE | -16.24 |
| Consensus MFE | -8.46 |
| Energy contribution | -9.17 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
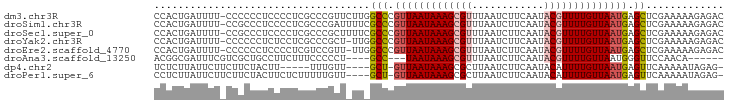
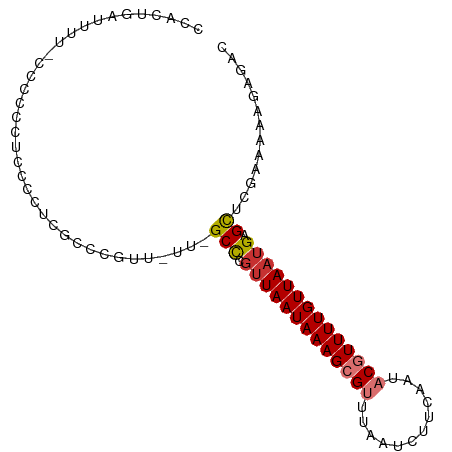
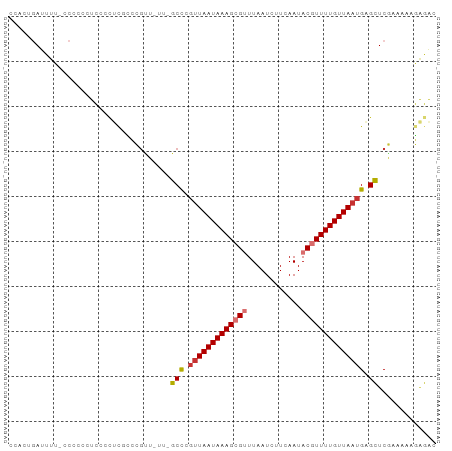
>dm3.chr3R 11119147 94 + 27905053 UUUGCCCACUGAUUUUCCCCCCUCCCCUCGCCCGUUCUUGGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAA ............(((((............(((.......))).((((((((((((((............)))))))))))))).....))))). ( -18.20, z-score = -3.30, R) >droSim1.chr3R 17170215 94 - 27517382 UUUGCCCACUGAUUUUCCGCCCUCCCCUCGCCCGAUUUUCGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAA ...................................(((((((.((((((((((((((............)))))))))))))).))..))))). ( -17.90, z-score = -2.94, R) >droSec1.super_0 10287793 94 - 21120651 UUUGCCCACUGAUUUUCCGCCCUCCCCUCGCCCGCUUUUCGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAA ............(((((.((.............)).....((.((((((((((((((............)))))))))))))).))..))))). ( -17.72, z-score = -2.86, R) >droYak2.chr3R 13517675 93 - 28832112 UUUGCCCACUGAUUUUCCCCCCUCUCCUCGCCCGCU-UUGGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAA ............(((((............(((....-..))).((((((((((((((............)))))))))))))).....))))). ( -18.70, z-score = -3.09, R) >droEre2.scaffold_4770 10642716 93 + 17746568 UUUGCCCACUGAUUUUCCCCCCUCCCCUCGUCCGUU-UUGGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAA ..................................((-(((((.((((((((((((((............)))))))))))))).)).))))).. ( -19.50, z-score = -3.89, R) >droAna3.scaffold_13250 3400663 87 - 3535662 UUUUCACGGCGAUUUCGUCGCUGCCUUCU------UUCCCCCUGCCUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGGGUUCCAACA- ......(((((((...)))))))......------....(((....(((((((((((............)))))))))))..)))........- ( -20.50, z-score = -3.11, R) >dp4.chr2 28045663 84 + 30794189 UUUGGUCUCUUAUUCUU---CUUCUACUU-----UUUGUUGCU-GUUAAUAAAGCGCUUAAUCUUCAAUACAUUUUGUUAAUGAGUUCAAAAA- ((((..(((........---.........-----......(((-........)))..........(((......))).....)))..))))..- ( -8.20, z-score = -0.25, R) >droPer1.super_6 3372113 89 + 6141320 UUUGGCCUCUUAUUCUU---CUUCUACUUCUCUUUUUGUUGCU-GUUAAUAAAGCGCUUAAUCUUCAAUACAUUUUGUUAAUGAGUUCAAAAA- ((((..(((........---....................(((-........)))..........(((......))).....)))..))))..- ( -9.20, z-score = -0.56, R) >consensus UUUGCCCACUGAUUUUCCCCCCUCCCCUCGCCCGUUUUUCGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAA ........................................((..(((((((((((((............)))))))))))))..))........ ( -8.46 = -9.17 + 0.72)



| Location | 11,119,152 – 11,119,246 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 76.23 |
| Shannon entropy | 0.47566 |
| G+C content | 0.40739 |
| Mean single sequence MFE | -18.36 |
| Consensus MFE | -8.93 |
| Energy contribution | -9.45 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.991057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11119152 94 + 27905053 CCACUGAUUUU-CCCCCCUCCCCUCGCCCGUUCUUGGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAAGAGAC ...........-..........(((.....(((..(((.((((((((((((((............)))))))))))))).))).)))...))).. ( -21.50, z-score = -3.63, R) >droSim1.chr3R 17170220 94 - 27517382 CCACUGAUUUU-CCGCCCUCCCCUCGCCCGAUUUUCGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAAGAGAC ...........-.....(((...((....))(((((((.((((((((((((((............)))))))))))))).))..))))).))).. ( -19.20, z-score = -2.67, R) >droSec1.super_0 10287798 94 - 21120651 CCACUGAUUUU-CCGCCCUCCCCUCGCCCGCUUUUCGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAAGAGAC ...........-..........((((....)(((((((.((((((((((((((............)))))))))))))).))..))))).))).. ( -19.20, z-score = -2.58, R) >droYak2.chr3R 13517680 93 - 28832112 CCACUGAUUUU-CCCCCCUCUCCUCGCCCGCU-UUGGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAAGAGAC ...........-.....((((..(((......-..(((.((((((((((((((............)))))))))))))).))))))...)))).. ( -21.00, z-score = -3.14, R) >droEre2.scaffold_4770 10642721 93 + 17746568 CCACUGAUUUU-CCCCCCUCCCCUCGUCCGUU-UUGGCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAAGAGAC ...........-..........(((.....((-(((((.((((((((((((((............)))))))))))))).)).)))))..))).. ( -22.70, z-score = -4.13, R) >droAna3.scaffold_13250 3400668 82 - 3535662 ACGGCGAUUUCGUCGCUGCCUUCUUUCCCCCU----GCC---UAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGGGUUCCAACA------ .(((((((...)))))))..........(((.----...---(((((((((((............)))))))))))..)))........------ ( -20.50, z-score = -3.15, R) >dp4.chr2 28045668 84 + 30794189 UCUCUUAUUCUUCUUCUACUU-----UUUGUU----GCU-GUUAAUAAAGCGCUUAAUCUUCAAUACAUUUUGUUAAUGAGUUCAAAAAUAGAG- ..............((((.((-----((((..----(((-((((((((((.(..............).)))))))))).))).)))))))))).- ( -11.54, z-score = -1.22, R) >droPer1.super_6 3372118 89 + 6141320 CCUCUUAUUCUUCUUCUACUUCUCUUUUUGUU----GCU-GUUAAUAAAGCGCUUAAUCUUCAAUACAUUUUGUUAAUGAGUUCAAAAAUAGAG- ....................(((.((((((..----(((-((((((((((.(..............).)))))))))).))).)))))).))).- ( -11.24, z-score = -1.23, R) >consensus CCACUGAUUUU_CCCCCCUCCCCUCGCCCGUU_UU_GCCCGUUAAUAAAGCGUUUAAUCUUCAAUACGUUUUGUUAAUGAGCUCGAAAAAGAGAC ....................................((..(((((((((((((............)))))))))))))..))............. ( -8.93 = -9.45 + 0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:57 2011