| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,112,855 – 11,112,945 |
| Length | 90 |
| Max. P | 0.997383 |

| Location | 11,112,855 – 11,112,945 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 71.47 |
| Shannon entropy | 0.53493 |
| G+C content | 0.26560 |
| Mean single sequence MFE | -16.23 |
| Consensus MFE | -9.31 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.997383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
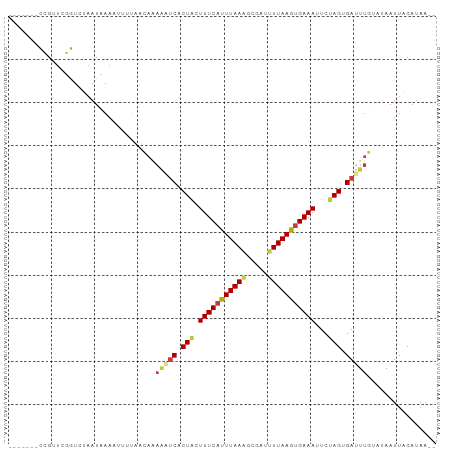
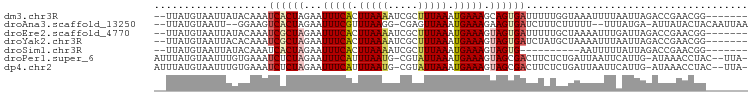
>dm3.chr3R 11112855 90 + 27905053 -------CCGUUCGGUCUAAUUAAAAUUUACCAAAAAUCACUGCUUUCAUUUAAAGCGAUUUUAAGUGAAAUUCUAGUGAUUUGUAUAAUUACAUAA-- -------......(((..(((....))).)))..(((((((((.((((((((((((...))))))))))))...)))))))))(((....)))....-- ( -17.90, z-score = -3.08, R) >droAna3.scaffold_13250 3394025 91 - 3535662 UUAAUUGUAGUAUAAU-UCAUAAA--AAAAAGAAAGAUCACUUCUUUCAUUUAACUCG-CCUUAAACGAAAUUCUAGUGACUUCC--AAUUACAUAA-- .....((((((..((.-((((...--.....((((((.....)))))).......(((-.......))).......)))).))..--.))))))...-- ( -11.70, z-score = -1.88, R) >droEre2.scaffold_4770 10636394 90 + 17746568 -------CCGUUCGGUCUAAUCAAAUUUUAGCAAAAAUCACUACUUUCAUUUAAAGCGAUUUUAAGUGAAAUUCUAGCGAUUUGUAUAAUUACAUAA-- -------.......(.((((.......)))))..(((((.(((.((((((((((((...))))))))))))...))).)))))(((....)))....-- ( -13.70, z-score = -1.36, R) >droYak2.chr3R 13511344 90 - 28832112 -------CCGUUCGGUCUAAUUAAAUUUUAGCAUAGAUCACUACUUUCAUUUAAAGCGAUUUUAAGUGAAAUUCUAGCGAUUUGUGUAAUUACAUAA-- -------((....))...............(((((((((.(((.((((((((((((...))))))))))))...))).)))))))))..........-- ( -19.60, z-score = -2.80, R) >droSim1.chr3R 17163423 80 - 27517382 -------CCGUUCGGUCUAAUAAAAAUU----------CACUACUUUCAUUUAAAGCGAUUUUAAGUGAAAUUCUAGUGAUUUGUAUAAUUACAUAA-- -------((....))............(----------(((((.((((((((((((...))))))))))))...))))))..((((....))))...-- ( -14.70, z-score = -2.41, R) >droPer1.super_6 3364423 94 + 6141320 -UAA--GUAGGUUUAU-CAAUGAAUUAAUCAGAGAAGUCGCUACUUUCAUUUAAUACG-CAUUAAAUGAAAUUCUAGAGAUUUCACAAAUUACAUAAAU -...--((((......-...(((.....)))..((((((.(((.(((((((((((...-.)))))))))))...))).)))))).....))))...... ( -18.00, z-score = -2.97, R) >dp4.chr2 28038020 94 + 30794189 -UAA--GUAGGUUUAU-CAAUGAAUUAAUCAGAGAAGUCGCUACUUUCAUUUAAUACG-CAUUAAAUGAAAUUCUAGAGAUUUCACAAAUUACAUAAAU -...--((((......-...(((.....)))..((((((.(((.(((((((((((...-.)))))))))))...))).)))))).....))))...... ( -18.00, z-score = -2.97, R) >consensus _______CCGUUCGGUCUAAUAAAAUUUUAACAAAAAUCACUACUUUCAUUUAAAGCGAUUUUAAGUGAAAUUCUAGUGAUUUGUAUAAUUACAUAA__ ..................................(((((.(((.(((((((((((.....)))))))))))...))).)))))................ ( -9.31 = -9.50 + 0.19)


| Location | 11,112,855 – 11,112,945 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 71.47 |
| Shannon entropy | 0.53493 |
| G+C content | 0.26560 |
| Mean single sequence MFE | -14.83 |
| Consensus MFE | -8.10 |
| Energy contribution | -8.39 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
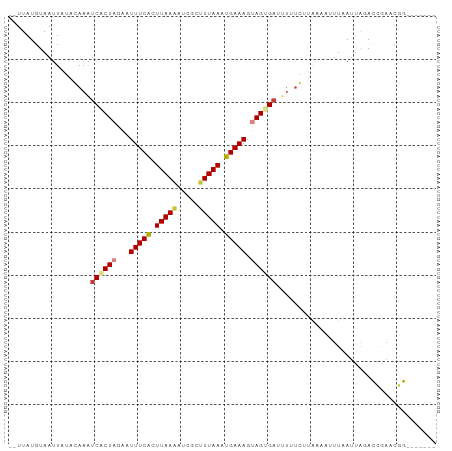
>dm3.chr3R 11112855 90 - 27905053 --UUAUGUAAUUAUACAAAUCACUAGAAUUUCACUUAAAAUCGCUUUAAAUGAAAGCAGUGAUUUUUGGUAAAUUUUAAUUAGACCGAACGG------- --...((((....))))(((((((....(((((.(((((.....))))).)))))..)))))))((((((.............))))))...------- ( -14.92, z-score = -1.65, R) >droAna3.scaffold_13250 3394025 91 + 3535662 --UUAUGUAAUU--GGAAGUCACUAGAAUUUCGUUUAAGG-CGAGUUAAAUGAAAGAAGUGAUCUUUCUUUUU--UUUAUGA-AUUAUACUACAAUUAA --(((((.((..--((((((((((....((((((((((..-....))))))))))..))))).)))))..)).--..)))))-................ ( -18.30, z-score = -1.79, R) >droEre2.scaffold_4770 10636394 90 - 17746568 --UUAUGUAAUUAUACAAAUCGCUAGAAUUUCACUUAAAAUCGCUUUAAAUGAAAGUAGUGAUUUUUGCUAAAAUUUGAUUAGACCGAACGG------- --....((((......(((((((((...(((((.(((((.....))))).))))).)))))))))))))...............((....))------- ( -13.50, z-score = -0.68, R) >droYak2.chr3R 13511344 90 + 28832112 --UUAUGUAAUUACACAAAUCGCUAGAAUUUCACUUAAAAUCGCUUUAAAUGAAAGUAGUGAUCUAUGCUAAAAUUUAAUUAGACCGAACGG------- --................(((((((...(((((.(((((.....))))).))))).))))))).....................((....))------- ( -11.40, z-score = -0.27, R) >droSim1.chr3R 17163423 80 + 27517382 --UUAUGUAAUUAUACAAAUCACUAGAAUUUCACUUAAAAUCGCUUUAAAUGAAAGUAGUG----------AAUUUUUAUUAGACCGAACGG------- --...((((....))))..((((((...(((((.(((((.....))))).))))).)))))----------)............((....))------- ( -12.30, z-score = -1.52, R) >droPer1.super_6 3364423 94 - 6141320 AUUUAUGUAAUUUGUGAAAUCUCUAGAAUUUCAUUUAAUG-CGUAUUAAAUGAAAGUAGCGACUUCUCUGAUUAAUUCAUUG-AUAAACCUAC--UUA- ...((((((.(..((((((((....).)))))))..).))-))))........((((((.....((..(((.....)))..)-).....))))--)).- ( -16.70, z-score = -2.11, R) >dp4.chr2 28038020 94 - 30794189 AUUUAUGUAAUUUGUGAAAUCUCUAGAAUUUCAUUUAAUG-CGUAUUAAAUGAAAGUAGCGACUUCUCUGAUUAAUUCAUUG-AUAAACCUAC--UUA- ...((((((.(..((((((((....).)))))))..).))-))))........((((((.....((..(((.....)))..)-).....))))--)).- ( -16.70, z-score = -2.11, R) >consensus __UUAUGUAAUUAUACAAAUCACUAGAAUUUCACUUAAAAUCGCUUUAAAUGAAAGUAGUGAUUUUUCUUAAAAUUUAAUUAGACCGAACGG_______ ...................((((((...(((((.(((((.....))))).))))).))))))..................................... ( -8.10 = -8.39 + 0.29)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:55 2011