| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,076,595 – 11,076,724 |
| Length | 129 |
| Max. P | 0.954796 |

| Location | 11,076,595 – 11,076,724 |
|---|---|
| Length | 129 |
| Sequences | 6 |
| Columns | 129 |
| Reading direction | reverse |
| Mean pairwise identity | 84.57 |
| Shannon entropy | 0.29345 |
| G+C content | 0.42000 |
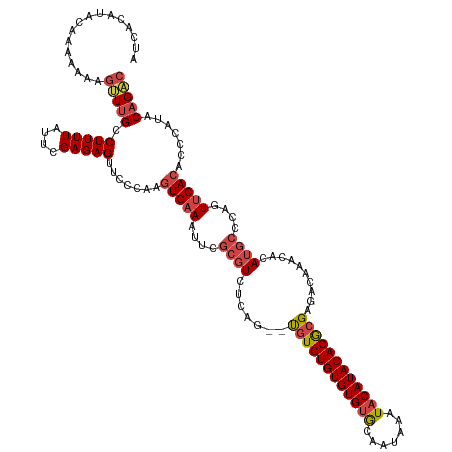
| Mean single sequence MFE | -32.75 |
| Consensus MFE | -20.18 |
| Energy contribution | -21.21 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
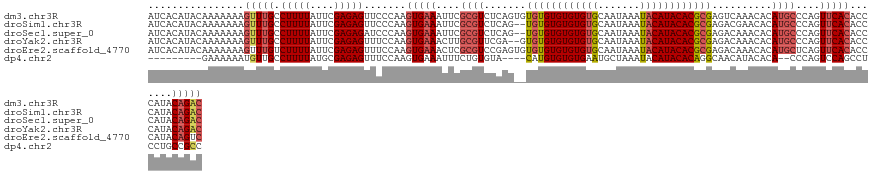
>dm3.chr3R 11076595 129 - 27905053 AUCACAUACAAAAAAAGUUUGCCUUUUAUUCGAGAGUUCCCAAGUGAAAUUCGCGUCUCAGUGUGUGUGUGUGUGCAAUAAAUACAUACACGCGAGUCAAACACAUGCCCAGUUCACACCCAUACAGAC ................(((((.(((((....))))).......(((((....(((((((.((((((((((((.........)))))))))))))))........))))....))))).......))))) ( -29.80, z-score = -1.67, R) >droSim1.chr3R 17128658 127 + 27517382 AUCACAUACAAAAAAAGUUUGCCUUUUAUUCGAGAGUUCCCAAGUGAAAUUCGCGUCUCAG--UGUGUGUGUGUGCAAUAAAUACAUACACGCGAGACGAACACAUGCCCAGUUCACACCCAUACAGAC ................(((((.(((((....))))).......(((((....(((((((.(--(((((((((((.......)))))))))))))))))(....)..))....))))).......))))) ( -33.90, z-score = -3.14, R) >droSec1.super_0 10245394 127 + 21120651 AUCACAUACAAAAAAAGUUUGCCUUUUAUUCGAGAGAUCCCAAGUGAAAUUCGCGUCUCAG--UGUGUGUGUGUGCAAUAAAUACAUACACGCGAGACAAACACAUGCCCAGUUCACACCCAUACAGAC ................(((((.(((((....))))).......(((((....(((((((.(--(((((((((((.......)))))))))))))))))........))....))))).......))))) ( -33.10, z-score = -3.30, R) >droYak2.chr3R 13474408 127 + 28832112 AUCACAUACAAAAAAAGUUUGCCUUUUAUUCGAGAGUUUCCAAGUGAAACUUGCGUUCGA--GUGUGUGUGUGUGCAAUAAAUACAUACACGCGAGACAAACACAUGCCCAGUUCACACCCAUACAGAC ................(((((.(((....(((((((((((.....)))))))....))))--((((((((((((.......))))))))))))))).)))))........................... ( -31.30, z-score = -2.00, R) >droEre2.scaffold_4770 10599864 129 - 17746568 AUCACAUACAAAAAAAGUUUGUCUUUUAUUCGAGAGUUUCCAAGUGAAACUCGCGUCCGAGUGUGUGUGUGUGUGCAAUAAAUACAUACACGCGAGACAAACACAUGCUCAGUUCACACCCAUACAGUC ................(((((((((..(((((.(((((((.....))))))).....)))))((((((((((((.......)))))))))))))))))))))........................... ( -38.40, z-score = -3.47, R) >dp4.chr2 27996591 114 - 30794189 ---------GAAAAAAUGUUGCCUUUUAUGCGAGAGUUUCCAAGUGAAAUUUCUGUGUA----CAUGUGUGUGAAUGCUAAAUACAUACACAGGCAACAUACACA--CCCAGUCCAGCCUCCUGCCGCC ---------......((((((((...((((((..((((((.....))))))..))))))----..((((((((.((.....)).)))))))))))))))).....--...................... ( -30.00, z-score = -2.45, R) >consensus AUCACAUACAAAAAAAGUUUGCCUUUUAUUCGAGAGUUCCCAAGUGAAAUUCGCGUCUCAG__UGUGUGUGUGUGCAAUAAAUACAUACACGCGAGACAAACACAUGCCCAGUUCACACCCAUACAGAC ................(((((.(((((....))))).......(((((....((((.......((((((((((((.......))))))))))))..........))))....))))).......))))) (-20.18 = -21.21 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:54 2011