
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,040,709 – 11,040,828 |
| Length | 119 |
| Max. P | 0.769113 |

| Location | 11,040,709 – 11,040,828 |
|---|---|
| Length | 119 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Shannon entropy | 0.48739 |
| G+C content | 0.46791 |
| Mean single sequence MFE | -37.49 |
| Consensus MFE | -12.79 |
| Energy contribution | -12.95 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.769113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11040709 119 + 27905053 UACUUGGAUAAUCGAAUGCUGGGCAUUGGUAGAAAGCCGGUGCUCAGCGGCAUUACAUCCAGCGAUAUGGAAUGACUGUCGACGUCCUUCAUGGAUACGACACCUAAAAGGUUAAGUAA- (((((((..........(((((((((((((.....)))))))))))))((.....((((((......))).)))..(((((..((((.....)))).))))))).......))))))).- ( -41.90, z-score = -3.55, R) >droSim1.chr3R 17092570 119 - 27517382 UAUUUGGACAAUCGAAUGCUGGGCAUUGGCAGAAAGCCGGUGCUCAGCGGCAUCACAUCCAGCGAUAUGGAAUGACUGUCGACGUCCUUCAUGGAUACGACACCUAAAAGGUUAAGUUA- ......(((..(((...(((((((((((((.....)))))))))))))(((((((..((((......)))).))).))))...((((.....)))).))).(((.....)))...))).- ( -42.70, z-score = -3.36, R) >droSec1.super_0 10209537 119 - 21120651 UAUUUGGACAAUCGAAUGCUGGGCAUUGGCAGAAAGCCGGUACUCAGCGGCAUUACAUCCAGCGAUAUGGAAUGACUGUCGACGUCCUUCAUGGAUACGACACCUAAAAGGUUAAGUAA- .....((((........((((((.((((((.....)))))).))))))((((...((((((......))).)))..))))...))))........(((...(((.....)))...))).- ( -36.30, z-score = -2.22, R) >droYak2.chr3R 13438424 119 - 28832112 UACUUGGAUAAUCGAAUGCUGGGCAUCGGUAGAAAGCCGGUGCUUAGCGGCAUCACAUCCAGCGAUAUGGAAUGACUGUCAACGUCCUUCAUGGAUACAACGCCUAAAAGGAUAAGUAA- ((((((......((...(((((((((((((.....)))))))))))))(((((((..((((......)))).))).))))..))(((((...((........))...))))))))))).- ( -42.00, z-score = -3.71, R) >droEre2.scaffold_4770 10562237 119 + 17746568 UACUUGGACAGUCGAAUGCUGGGCAUUGGUAGAAAGCCGGUGCUUAGCGGCAUCACAUCCAGCGAUAUGGAAUGACUAUCAACAUCCUUCAUGGAUACGACCCCUAAAAGGUUGAGUAA- ((((..((.((((((.((((((((((((((.....))))))))))))))...))...((((......))))..)))).))...((((.....)))).(((((.......))))))))).- ( -41.20, z-score = -3.05, R) >droAna3.scaffold_13250 3321688 116 - 3535662 UACUUAGAUAGUCGGAUGCUAGGCAUGGGUAGAAAGCCCGUGCUUAGCGGCAUAACAUCCAGCGAUAUGGAGUGACUGUCGACGUCUUUCAUGGACACAACACCUGAAAUAAUUGA---- ......(((((((...((((((((((((((.....)))))))))))))).....((.((((......)))))))))))))...((((.....))))....................---- ( -42.70, z-score = -4.11, R) >dp4.chr2 26273055 120 + 30794189 UAUUUGGACAGACGUAUGCUGGGCAUGGGCAGGAAGCCAGUGCUCAAGGGCAUCACAUCCAGGGAUAUGGAAUGGCUGUCCACGUCUUUCAUGGAGACGACACCUAAAAGGAUUUAGAGA ....(((((((.((((((((((((((.(((.....))).))))))...))))).)).((((......))))...))))))))((((((.....))))))....(((((....)))))... ( -42.20, z-score = -2.73, R) >droPer1.super_6 1602915 120 + 6141320 UAUUUGGACAGACGUAUGCUGGGCAUGGGCAGGAAGCCAGUGCUCAAGGGCAUCACAUCCAGGGAUAUGGAAUGGCUGUCCACGUCUUUCAUGGAGACGACACCUAAAAGGAUUUAGGGA ....(((((((.((((((((((((((.(((.....))).))))))...))))).)).((((......))))...))))))))((((((.....))))))...((((((....)))))).. ( -45.90, z-score = -3.65, R) >droWil1.scaffold_181089 5196695 114 + 12369635 UAUUUUGAUAAUCGUAUACUUGGCAUGGGUAGGAAUCCAGUGCUUAAAGGCAUAACAUCGAGAGAUAUUGAAUGACUAUCAACGUCUUUCAUUGAAACAACUCCUGAA--AAAUGA---- ....((((((.((((...((((((((((((....))))).))))..)))...(((.(((....))).))).)))).))))))(((.(((((..((......)).))))--).))).---- ( -22.90, z-score = -0.63, R) >droVir3.scaffold_13047 4166448 116 + 19223366 UACUUGGACAGGCGUAUGCUGGGCAUAGGCAGAAAGCCAGUACUCAGGGGCAUAACGUCCAGCGAUAUCGAAUGACAGUCCACAUCCUUCAUGGAUACGACACCUACAAGAUUGUU---- ..((((..((..((((((((((((...(((.....))).(..(....)..).....))))))).))).))..))...(((...((((.....))))..))).....))))......---- ( -33.10, z-score = -1.29, R) >droMoj3.scaffold_6540 21150252 116 + 34148556 UACUUAGACAGUCGUAUACUGGGCAUGGGCAGAAAGCCAGUACUUAGUGGCAUAACAUCCAGCGAUAUCGAAUGACAGUCCACAUCCUUCAUGGACACGACGCCUGAAAUAUUGUU---- ...((((...(((((...((((((....)).....((((.(....).)))).......))))...............(((((.........))))))))))..)))).........---- ( -26.40, z-score = 0.09, R) >droGri2.scaffold_14906 805662 111 + 14172833 UAUUUCGACAGUCGUAUGCUGGGCAUAGGCAAAAAGCCAGUGCUCAGCGGCAUAACAUCCAGCGAUAUAGAAUGGCUAUCCACAUCCCUCAUGGAUACGACACCUGCAAGA--------- ..........((((((((((((((((.(((.....))).))))))))))........((((..((.......(((....)))......)).))))))))))..........--------- ( -34.62, z-score = -2.15, R) >anoGam1.chr2R 44656872 117 - 62725911 UAUUUCGACAAACGAAUAUUGGGCAGUGGUAAAAAGCCGGCCGUCAGCGGAAGCACAUCUAGCGUGAUGCUGUGGCUUUCCACCUCUUCCAUCGAGAUGACUCCUGCAAUUGAGUAA--- (((((((((((.......)))((.((.(((..((((((..(((....)))..(((((((......)))).)))))))))..))).)).)).))))))))((((........))))..--- ( -35.40, z-score = -1.19, R) >consensus UAUUUGGACAGUCGAAUGCUGGGCAUGGGCAGAAAGCCAGUGCUCAGCGGCAUCACAUCCAGCGAUAUGGAAUGACUGUCCACGUCCUUCAUGGAUACGACACCUAAAAGGUUAAA____ ...................((((....(((((...(((..........)))....((((((......))).))).)))))...((((.....))))......)))).............. (-12.79 = -12.95 + 0.16)
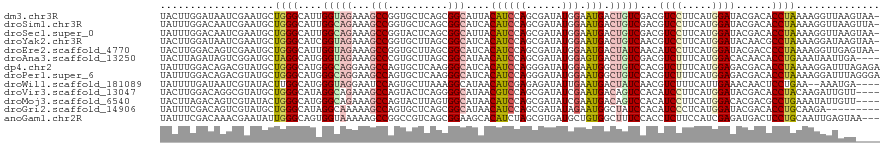


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:51 2011