| Sequence ID | dm3.chr3R |
|---|---|
| Location | 11,005,915 – 11,006,029 |
| Length | 114 |
| Max. P | 0.991190 |

| Location | 11,005,915 – 11,006,029 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.79 |
| Shannon entropy | 0.21338 |
| G+C content | 0.34476 |
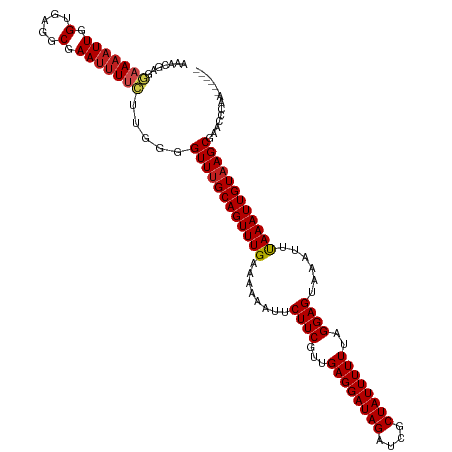
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -23.60 |
| Energy contribution | -23.71 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 11005915 114 - 27905053 AAACGAGGAAAAUUGGUGAGGCGAAUUUUCUUGGGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAAUUUAAAUUGUAAGCGAACCCGA------ .....(((((((((.(.....).)))))))))(((((((((((((((((......((((...((((((((....))))))))..))))....))))))))))))))...)))..------ ( -33.80, z-score = -3.33, R) >droGri2.scaffold_14830 3542604 118 - 6267026 AAACGAGUAAAAUUGGUGAGGCGAAUAUCGA-AUCGUUGGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAA-GCAAAUUGUAAGCCAAGCAACAAAUAG ......((....(((((((..((.....)).-.))....((((((((........((((...((((((((....))))))))..))))....-.))))))))..)))))...))...... ( -25.52, z-score = -1.30, R) >droMoj3.scaffold_6540 3663578 111 - 34148556 AAACGAGUAAAAUUGGUGAGGCGAAAUUUGA-AUGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAA-GCAAAUUGUAAGCCAUAUCA------- ...(((......))).............(((-(((((((((((((((........((((...((((((((....))))))))..))))....-.))))))))))))))).)))------- ( -28.62, z-score = -2.94, R) >droVir3.scaffold_13047 7728419 118 - 19223366 AAACGAGUAAAAUUGGUGAGGCAAAUUUCGA-AUGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAA-GCAAAUUGUAAGCUAAGCAAAAAAAAA ...((((.....(((......)))..)))).-.((((((((((((((........((((...((((((((....))))))))..))))....-.))))))))))))))............ ( -25.92, z-score = -1.84, R) >droWil1.scaffold_181130 9401049 113 + 16660200 AAACGAGGAAAAUUGGUGAGCCAAAUUUUCAUUUGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAA-UUAAAUUGUAAGCCGAUAAAU------ ......(((((.((((....)))).)))))..((((((((((((((((.......((((...((((((((....))))))))..))))....-)))))))))))))))).....------ ( -33.20, z-score = -4.37, R) >droPer1.super_3 3739852 114 - 7375914 AAACGAGUAAAAUUGGUGAGGCGAAUUUUCUUGGGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAUUUUAAAUUGUAAGCCAACUGAA------ ...((((.((((((.(.....).)))))))))).((((((((((((((((.....((((...((((((((....))))))))..))))...)))))))))))))))).......------ ( -34.30, z-score = -4.15, R) >dp4.chr2 20989231 114 - 30794189 AAACGAGUAAAAUUGGUGAGGCGAAUUUUCUUGGGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAUUUUAAAUUGUAAGCCAACUGAA------ ...((((.((((((.(.....).)))))))))).((((((((((((((((.....((((...((((((((....))))))))..))))...)))))))))))))))).......------ ( -34.30, z-score = -4.15, R) >droAna3.scaffold_13340 6089379 114 - 23697760 AAACGAGGAAAAUUGGUGAGGCGAAUUUUCAUGGGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAUUUUAAAUUGUAAGCGAAACCGA------ .......(((((((.(.....).))))))).(((.(((((((((((((((.....((((...((((((((....))))))))..))))...)))))))))))))))....))).------ ( -31.30, z-score = -3.05, R) >droEre2.scaffold_4770 10527406 114 - 17746568 AAACGAGGAAAAUUGGUGAGGCGAAUUUUCUUGGGGUUUGCAGUUUGAGAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAAUUUAAAUUGUAAGCGAACCCGA------ .....(((((((((.(.....).)))))))))(((((((((((((((((......((((...((((((((....))))))))..))))....))))))))))))))...)))..------ ( -33.50, z-score = -3.12, R) >droYak2.chr3R 13403207 114 + 28832112 AAACGAGGAAAAUUGGUGAGGCGAAUUUUCUUGGGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAAUUUAAAUUGUAAGCGAACCCGA------ .....(((((((((.(.....).)))))))))(((((((((((((((((......((((...((((((((....))))))))..))))....))))))))))))))...)))..------ ( -33.80, z-score = -3.33, R) >droSec1.super_0 10174624 114 + 21120651 AAACGAGGAAAAUUGGUGAGGCGAAUUUUCUUGGGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAAUUUAAAUUGUAAGCGAACCCGA------ .....(((((((((.(.....).)))))))))(((((((((((((((((......((((...((((((((....))))))))..))))....))))))))))))))...)))..------ ( -33.80, z-score = -3.33, R) >droSim1.chr3R 17057287 114 + 27517382 AAACGAGGAAAAUUGGUGAGGCGAAUUUUCUUGGGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAAUUUAAAUUGUAAGCGAACCCGA------ .....(((((((((.(.....).)))))))))(((((((((((((((((......((((...((((((((....))))))))..))))....))))))))))))))...)))..------ ( -33.80, z-score = -3.33, R) >consensus AAACGAGGAAAAUUGGUGAGGCGAAUUUUCUUGGGGUUUGCAGUUUGAAAAAAUUCUUCGUUGAGGAUAGAUCGCUAUUUUUUAGGAGUAAAUUUAAAUUGUAAGCGAACCCAA______ .......(((((((.(.....).))))))).....((((((((((((........((((...((((((((....))))))))..))))......)))))))))))).............. (-23.60 = -23.71 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:47 2011