
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,997,015 – 10,997,116 |
| Length | 101 |
| Max. P | 0.861243 |

| Location | 10,997,015 – 10,997,116 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.25 |
| Shannon entropy | 0.48415 |
| G+C content | 0.36577 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -12.38 |
| Energy contribution | -12.50 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.861243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
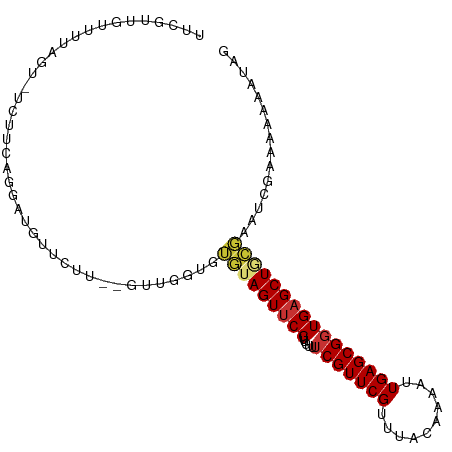
>dm3.chr3R 10997015 101 + 27905053 UUCGUUGUUUUAGU-UCUUCAGGAUGUUAUUUAGUUGGUGUGUAGUUCGUUGUUUCGUUCGUUUACAAAAUUGAGCGGUGAGCUGCGAAUCGGAAAUAAUAG ...((((((((.((-((....))))..........((((.(((((((((..((((.(((.........))).))))..))))))))).)))))))))))).. ( -22.50, z-score = -1.55, R) >droGri2.scaffold_14830 3533771 81 + 6267026 UUCGUUGUUGUAAU-------UGUUGUU--------GCUUUAAAGUUCGUUUU--CGUUCGUUUACAAAAUUGAGCGGUGUGCUGUCAAAAGUGAUAG---- ..............-------...(((.--------.((((..(((.((...(--((((((..........))))))))).)))....))))..))).---- ( -13.50, z-score = -0.34, R) >droWil1.scaffold_181130 9390601 93 - 16660200 UUCGUUGUUGGUAU-------UCUUGUUUUCU-GAUGUUGUUUAGUUCGUUUUUUCGUUCGUUUACAAAAUUGAGCGGUGAGCUGUGAAUAUAGAAUAUUA- ........((((((-------((((((((...-.........(((((((.....(((((((..........)))))))))))))).))))).)))))))))- ( -16.72, z-score = -1.00, R) >droAna3.scaffold_13340 6080822 96 + 23697760 UUCGUCGUUGU----UUUUCAGGAUGUUCUU--GUUGUUGAAAAGUUCGUUGUUUCGUUCGUUUACAAAAUUGAGCGGUGAGCUGUUGGGAAUAUAAAUUAA ..((((.(((.----....)))))))....(--(((.(..(..((((((..((((.(((.........))).))))..)))))).)..).))))........ ( -15.90, z-score = 0.30, R) >droEre2.scaffold_4770 10518583 98 + 17746568 UUCGUUGUUUUUGU-UCUACAGGAUGUUCUU--UUUGGU-UGUAGUUCGUUGUUUCGUUCGUUUACAAAAUUGAGCGGUGAGCUACGGGUCGAAAAAUAUGG ..(((..((((((.-.((.(((((......)--))))..-.((((((((..((((.(((.........))).))))..))))))))))..))))))..))). ( -22.20, z-score = -1.41, R) >droYak2.chr3R 13394132 100 - 28832112 UUCGUUGUUUUUGGAUGUUUAGGAUGUUCUU--GUUGGUGUGUAGUUCGUUGUUUCGUUCGUUUACAAAAUUGAGCGGUGAGCUACGGAUCGAAAAAUGUAG ..((((......(((((((...)))))))..--.(((((.(((((((((..((((.(((.........))).))))..))))))))).)))))..))))... ( -22.70, z-score = -1.53, R) >droSec1.super_0 10165860 99 - 21120651 UUCGUUGUUUUAGU-UCUUCAGGAUGUUUUU--GUUGGUUUGUAGUUCGUUGUUUCGUUCGUUUACAAAAUUGAGCGGUGAGCUGCGAAUCGGAUAAAAUAG .....(((((((((-((....))))......--.(((((((((((((((..((((.(((.........))).))))..))))))))))))))).))))))). ( -25.50, z-score = -2.85, R) >droSim1.chr3R 17047650 99 - 27517382 UUCUUUGUUUUAGU-UCUUCAGGAUGUUUUU--GUUGGUUUGUAGUUCGUUGUUUCGUUCGUUUACAAAAUUGAGCGGUGAGCUGCGAAUCGGAUAAAAUAG .....(((((((((-((....))))......--.(((((((((((((((..((((.(((.........))).))))..))))))))))))))).))))))). ( -25.50, z-score = -2.96, R) >consensus UUCGUUGUUUUAGU_UCUUCAGGAUGUUCUU__GUUGGUGUGUAGUUCGUUGUUUCGUUCGUUUACAAAAUUGAGCGGUGAGCUGCGAAUCGAAAAAAAUAG ........................................(((((((((.....(((((((..........))))))))))))))))............... (-12.38 = -12.50 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:46 2011