
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,987,297 – 10,987,387 |
| Length | 90 |
| Max. P | 0.845404 |

| Location | 10,987,297 – 10,987,387 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.80 |
| Shannon entropy | 0.42307 |
| G+C content | 0.37123 |
| Mean single sequence MFE | -19.07 |
| Consensus MFE | -8.94 |
| Energy contribution | -9.05 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
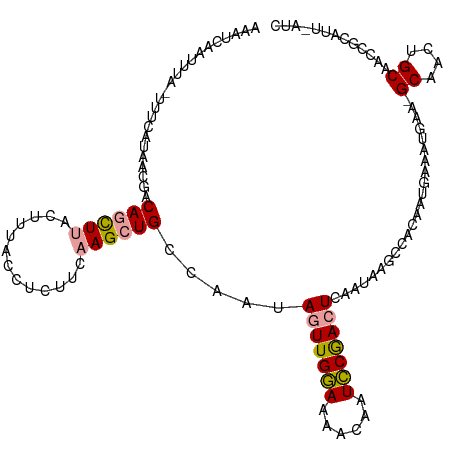
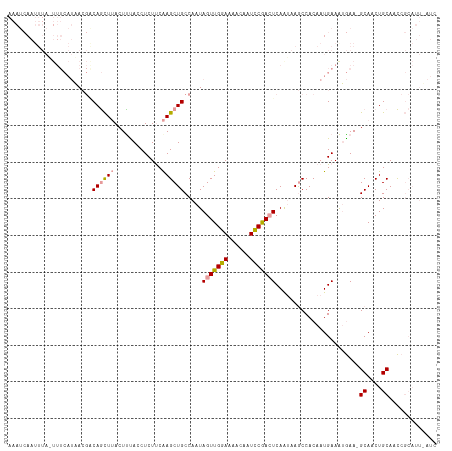
>dm3.chr3R 10987297 90 + 27905053 -----------------AAACCUCAAUUAACUUUACCUCUUCGAGCUGCCAAUAGUUGAAAAACAAUUCGAAUCAAUAAGCCACAAUGAAAUAAA-GCAACUGCAAUC--------- -----------------.............(((((..((.....(((........(((((......))))).......)))......))..))))-)...........--------- ( -7.46, z-score = 0.05, R) >droEre2.scaffold_4770 10513575 117 + 17746568 AAAUCAAUUUAAUUUCAUUAGGACAGCUUACUCUACCUCUCCAAGCUGCCAAUAGUUGGAGAACAAUCCAACUCAAUAAACCACAAUGAAGAGAACGCAACUGCAACCGCAUUAAUC ............(((((((.((.((((((.............))))))))...(((((((......)))))))...........))))))).....((....))............. ( -23.52, z-score = -3.32, R) >droYak2.chr3R 13389133 117 - 28832112 AAAUCAAUUUAUUUUCAUAACGGCAGCUUACUUCACCUCUUCAAGCUGCCAAUAGUUGGAAAACAAUCCGACUCAGAAAGCCACAAUGAAAUGAAAGCAACUGCAACCGCAUUUAUC .......(((((((.(((...((((((((.............))))))))...(((((((......)))))))............))))))))))......(((....)))...... ( -26.22, z-score = -3.38, R) >consensus AAAUCAAUUUA_UUUCAUAACGACAGCUUACUUUACCUCUUCAAGCUGCCAAUAGUUGGAAAACAAUCCGACUCAAUAAGCCACAAUGAAAUGAA_GCAACUGCAACCGCAUU_AUC .......................((((((.............)))))).....(((((((......))))))).......................((....))............. ( -8.94 = -9.05 + 0.12)

| Location | 10,987,297 – 10,987,387 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.80 |
| Shannon entropy | 0.42307 |
| G+C content | 0.37123 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -18.63 |
| Energy contribution | -17.43 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
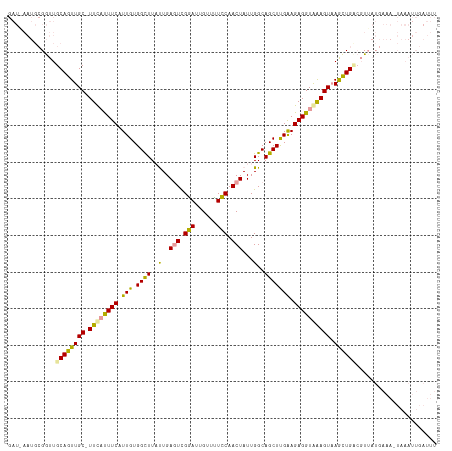
>dm3.chr3R 10987297 90 - 27905053 ---------GAUUGCAGUUGC-UUUAUUUCAUUGUGGCUUAUUGAUUCGAAUUGUUUUUCAACUAUUGGCAGCUCGAAGAGGUAAAGUUAAUUGAGGUUU----------------- ---------(((..(((((((-((((((((.(((.((((..(..((..(((......)))....))..).))))))).))))))))).))))))..))).----------------- ( -20.30, z-score = -1.10, R) >droEre2.scaffold_4770 10513575 117 - 17746568 GAUUAAUGCGGUUGCAGUUGCGUUCUCUUCAUUGUGGUUUAUUGAGUUGGAUUGUUCUCCAACUAUUGGCAGCUUGGAGAGGUAGAGUAAGCUGUCCUAAUGAAAUUAAAUUGAUUU (((((((..((((((((((((..((((((((..((.(((.....(((((((......)))))))...))).)).))))))))....)).))))))........))))..))))))). ( -29.70, z-score = -1.09, R) >droYak2.chr3R 13389133 117 + 28832112 GAUAAAUGCGGUUGCAGUUGCUUUCAUUUCAUUGUGGCUUUCUGAGUCGGAUUGUUUUCCAACUAUUGGCAGCUUGAAGAGGUGAAGUAAGCUGCCGUUAUGAAAAUAAAUUGAUUU .......(((((.((..((((((.((((((.....((((.((..(((.(((......))).)))...)).))))....)))))))))))))).)))))(((....)))......... ( -30.60, z-score = -1.12, R) >consensus GAU_AAUGCGGUUGCAGUUGC_UUCAUUUCAUUGUGGCUUAUUGAGUCGGAUUGUUUUCCAACUAUUGGCAGCUUGAAGAGGUAAAGUAAGCUGACGUUAUGAAA_UAAAUUGAUUU .............((((((((.((((((((.(((.((((..(..(((.(((......))).)))...)..))))))).)))))))))).))))))...................... (-18.63 = -17.43 + -1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:45 2011