
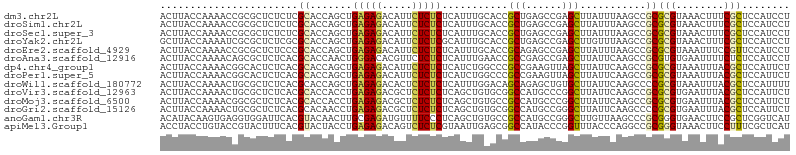
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,495,506 – 7,495,611 |
| Length | 105 |
| Max. P | 0.943252 |

| Location | 7,495,506 – 7,495,611 |
|---|---|
| Length | 105 |
| Sequences | 14 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Shannon entropy | 0.44217 |
| G+C content | 0.52993 |
| Mean single sequence MFE | -30.66 |
| Consensus MFE | -11.43 |
| Energy contribution | -11.32 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
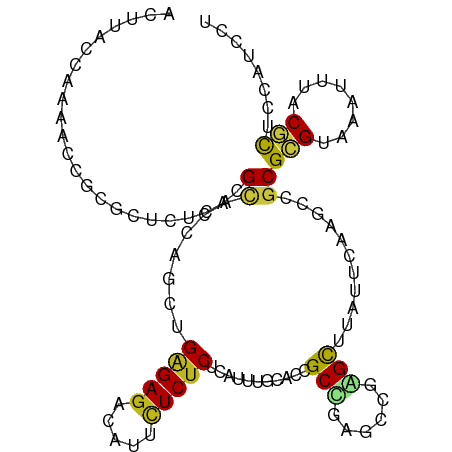
>dm3.chr2L 7495506 105 + 23011544 ACUUACCAAAACCGCGCUCUCUCGCACCAGCUGAGAGACAUUCUCUCUCAUUUGCACCGCUGAGCCGAGCUUAUUUAAGCCGCGCGUAAACUUUCGCUCCAUCCU .............((((((...((...(((.(((((((.....))))))).)))...))..)))).(.((((....)))))))(((........)))........ ( -25.70, z-score = -2.05, R) >droSim1.chr2L 7288126 105 + 22036055 ACUUACCAAAACCGCGCUCUCUCGCACCAGCUGAGAGACAUUCUCUCUCAUUUGCACCGCUGAGCCGAGCUUAUUUAAGCCGCGCGUAAACUUUCGCUCCAUCCU .............((((((...((...(((.(((((((.....))))))).)))...))..)))).(.((((....)))))))(((........)))........ ( -25.70, z-score = -2.05, R) >droSec1.super_3 3011895 105 + 7220098 ACUUACCAAAACCGCGCUCUCUCGCACCAGCUGAGAGACAUUCUCUCUCAUUUGCACCGCUGAGCCGAGCUUAUUUAAGCCGCGCGUAAACUUUCGCUCCAUCCU .............((((((...((...(((.(((((((.....))))))).)))...))..)))).(.((((....)))))))(((........)))........ ( -25.70, z-score = -2.05, R) >droYak2.chr2L 16923308 105 - 22324452 GCUUACCAAAAUCGCGCUCUCGCGCACCAGCUGAGAGACAUUCUCUCGCAUUUGCACCGCUGAGCCGAGCUUGUUUAAGCCGCGCGUAAACUUUCGCUCCAUCCU (((..........((((....))))..((((.(((((.....)))))((....))...))))))).((((..(((((.((...)).)))))....))))...... ( -32.00, z-score = -1.93, R) >droEre2.scaffold_4929 16416033 105 + 26641161 ACUUACCAAAACCGCGCUCUCCCGCACCAGCUGAGAGACAUUCUCUCUCAUUUGCACCGCAGAGCCGAGCUUAUUUAAGCCGCGCGUAAAUUUCCGUUCCAUCCU ..........((((((((((..((...(((.(((((((.....))))))).)))...)).))))).(.((((....)))))))).)).................. ( -25.50, z-score = -2.75, R) >droAna3.scaffold_12916 793508 105 - 16180835 ACUUACCAAAACAGCGCUCUCACGCACCAACUGGGACACGUUCUCUCUCAUUUGAACCGCCGAGCCGAGCUUAUUCAAGCCGCGUGUGAAUUUUCUCUCCAUCCU .................((.(((((.....((.((....((((..........))))..)).))..(.((((....)))))))))).))................ ( -19.60, z-score = -0.25, R) >dp4.chr4_group1 4275363 105 + 5278887 ACUUACCAAAACGGCACUCUCACGCACCAGCUGAGAGACAUUCUCUCUCAUCUGGCCCGCCGAAGUUAGCUUAUUCAAGCCGCGCGUAAAUUUACGCUCCAUUCU ...........((((.......((..((((.(((((((.....))))))).))))..))...(((....)))......)))).(((((....)))))........ ( -29.80, z-score = -3.72, R) >droPer1.super_5 5877989 105 - 6813705 ACUUACCAAAACGGCACUCUCACGCACCAGCUGAGAGACAUUCUCUCUCAUCUGGCCCGCCGAAGUUAGCUUAUUCAAGCCGCGCGUAAAUUUACGCUCCAUUCU ...........((((.......((..((((.(((((((.....))))))).))))..))...(((....)))......)))).(((((....)))))........ ( -29.80, z-score = -3.72, R) >droWil1.scaffold_180772 3771316 105 - 8906247 ACUUACCAAAACUGCGCUCUCACGCACCAGCUGAGAGACACUCUCUCUCAUUUGGACAGCAGAGCUGUGCUUAUUCAAGCCCCGCGUAAAUUUACGCUCCAUUUU ............((((......))))((((.(((((((.....))))))).))))(((((...)))))((((....))))...(((((....)))))........ ( -29.80, z-score = -3.39, R) >droVir3.scaffold_12963 4885308 105 - 20206255 ACUUACCAAAACUGCGCUCUCACGCACCACCUGAGAGACGCUCUCUCUCAGCUGUGCGGCCAUGCCCGGCUUAUUCAAGCCGCGCGUGAAUUUACGCUCCAUUCU .............(((......(((((...((((((((.....))))))))..)))))..(((((.((((((....)))))).)))))......)))........ ( -39.10, z-score = -5.13, R) >droMoj3.scaffold_6500 14054763 105 - 32352404 ACUUACCAAAACGGCGCUCUCACGCACCACCUGAGAGACGCUCUCUCUCAGCUGUGCCGCCAUGCCCGGCUUAUUCAAGCCGCGCGUGAAUUUACGCUCCAUUCU ............((.((......((((...((((((((.....))))))))..))))...(((((.((((((....)))))).))))).......)).))..... ( -38.30, z-score = -4.89, R) >droGri2.scaffold_15126 4105310 105 + 8399593 ACUUACCAAAACUGCGCUCUCACGCACAACCUGAGAGACGCUCUCUCUCAGCUGUGCGGCCAUGCCGGGCUUAUUCAAGCCCCGCGUGAAUUUACGCUCCAUUCU .............(((......((((((..((((((((.....)))))))).))))))..(((((.((((((....)))))).)))))......)))........ ( -41.50, z-score = -5.93, R) >anoGam1.chr3R 12490126 105 + 53272125 ACAUACAAGUGAGGUGGAUUCACGUACAACUUGCGAGAUGUUUUCCCUCAGCUGUGCCGCCAUGCCGGGCUUGUUAAGCCCGCGGGUGAACUUCCGCUCGGUCAU ...((((..(((((..((.((.((((.....)))).))...))..)))))..)))).....((((((((((.....)))))(((((......)))))..)).))) ( -35.50, z-score = -0.84, R) >apiMel3.Group1 2407560 105 - 25854376 ACCUACCUGUACCGUACUUUCACGUACUACCUGAGAGACAGUCUCUCGUAAUUGAGCGGCCAUACCCGGUUUACCCAGGCCGCGGGUAAACUUCCUUUCGCUCAU .........(((.((((......)))).....(((((.....))))))))..(((((((........(((((((((.......))))))))).....))))))). ( -31.22, z-score = -2.37, R) >consensus ACUUACCAAAACCGCGCUCUCACGCACCAGCUGAGAGACAUUCUCUCUCAUUUGCACCGCCGAGCCGAGCUUAUUCAAGCCGCGCGUAAAUUUACGCUCCAUCCU .......................((.......(((((.....)))))...........(((......)))...........))(((........)))........ (-11.43 = -11.32 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:23:48 2011