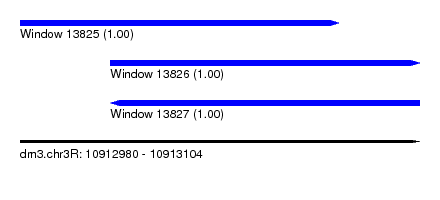
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,912,980 – 10,913,104 |
| Length | 124 |
| Max. P | 0.997063 |

| Location | 10,912,980 – 10,913,079 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 76.53 |
| Shannon entropy | 0.34578 |
| G+C content | 0.50561 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -14.12 |
| Energy contribution | -15.88 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.995745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10912980 99 + 27905053 CCCCGCCUGGGGAAACCCCCCAUGUCCCCCCCCCCUUCCUUUAACCGC-----ACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACAC .......(((((.....)))))((((....................((-----....))..................(((((((((....))))))))))))). ( -24.60, z-score = -4.48, R) >droSim1.chr3R 16971427 88 - 27517382 ----------------CCCCGCCGGGGCAAACCCCCCCUUUGAACCACCGCACACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACGC ----------------.......((((....))))..............((......))................(((((((((((....)))))))))))... ( -20.90, z-score = -2.62, R) >droSec1.super_0 10097754 88 - 21120651 ----------------CCCCGCCGGGGGAAACCCCCCCUUUGAACCGCCGCACACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACGC ----------------.......((((....))))...........((.((......))................(((((((((((....))))))))))).)) ( -25.40, z-score = -3.14, R) >droEre2.scaffold_4770 10448081 83 + 17746568 ----------------CCCCGCCGCCGGAGAAACCCCCUUGGAGCCGC-----ACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACGC ----------------....((.(((.(((.......))).).)).))-----......................(((((((((((....)))))))))))... ( -20.80, z-score = -2.52, R) >consensus ________________CCCCGCCGGCGCAAACCCCCCCUUUGAACCGC_____ACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACGC .......................(((((.....))))).....................................(((((((((((....)))))))))))... (-14.12 = -15.88 + 1.75)

| Location | 10,913,008 – 10,913,104 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.37 |
| Shannon entropy | 0.49352 |
| G+C content | 0.41610 |
| Mean single sequence MFE | -20.53 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.40 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.995843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10913008 96 + 27905053 CCCCCCCUUCCUUUAAC-CGCACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACACUAU-AUGCAAGUAGACGACAGCCCAG------------- .................-.((....((((((((......))).(((((((((((....))))))))))).....)-))))..((.....)).))....------------- ( -17.00, z-score = -3.45, R) >droSim1.chr3R 16971448 92 - 27517382 -----CUUUGAACCACCGCACACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACGCCAU-AUGCAAGUAGACAACAGCCCAG------------- -----............((......((((((((......))).(((((((((((....))))))))))).....)-))))..((.....)).))....------------- ( -16.90, z-score = -2.26, R) >droSec1.super_0 10097775 92 - 21120651 -----CUUUGAACCGCCGCACACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACGCCAU-AUGCAAGUAGACGACAGCCCAG------------- -----.........((.((......))....((((((......(((((((((((....))))))))))).((...-..)).)))))).....))....------------- ( -17.30, z-score = -1.55, R) >droYak2.chr3R 13323620 87 - 28832112 ----------CUUGGAGCCGCACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACGCUAU-AUGCAAGUAGACAGCCGACAGC------------- ----------.((((....((....))....((((((......(((((((((((....))))))))))).((...-..)).))))))...))))....------------- ( -20.20, z-score = -1.62, R) >droEre2.scaffold_4770 10448102 80 + 17746568 ----------CUUGGAGCCGCACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACGCCAU-AUGCAAGUAGACAGC-------------------- ----------((((.(...((....))................(((((((((((....)))))))))))......-.).))))........-------------------- ( -16.90, z-score = -1.19, R) >dp4.chr2 20891719 100 + 30794189 -----------CUCCGAGCACACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGAGUCGAUGAUAUGCACAAGCAGUAGACAAACGCUUUGCUUUGG -----------..(((((((.....(((((.....((((...((((((((((((....))))))))))))...)))))))))..((((.((......))))))))).)))) ( -27.70, z-score = -3.07, R) >droPer1.super_3 3643660 100 + 7375914 -----------CUCCGAGCACACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGAGUCGAUGAUAUGCACAAGCAGUAGACAAACGCUUUGCUUUGG -----------..(((((((.....(((((.....((((...((((((((((((....))))))))))))...)))))))))..((((.((......))))))))).)))) ( -27.70, z-score = -3.07, R) >consensus __________CUUCGACGCACACACGCAUAAUUUAUUAUAAUUUUGACAUUGAACGAGUUCAAUGUCGACGCCAU_AUGCAAGUAGACAGCAGACAAG_____________ .........................((((((((......))))(((((((((((....))))))))))).......))))............................... (-13.32 = -13.40 + 0.08)


| Location | 10,913,008 – 10,913,104 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.37 |
| Shannon entropy | 0.49352 |
| G+C content | 0.41610 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -15.11 |
| Energy contribution | -15.01 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.997063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10913008 96 - 27905053 -------------CUGGGCUGUCGUCUACUUGCAU-AUAGUGUCGACAUUGAACUCGUUCAAUGUCAAAAUUAUAAUAAAUUAUGCGUGUGCG-GUUAAAGGAAGGGGGGG -------------.....((.((.((((((.((((-((......(((((((((....))))))))).....(((((....))))).)))))))-))....))).)).)).. ( -22.60, z-score = -1.27, R) >droSim1.chr3R 16971448 92 + 27517382 -------------CUGGGCUGUUGUCUACUUGCAU-AUGGCGUCGACAUUGAACUCGUUCAAUGUCAAAAUUAUAAUAAAUUAUGCGUGUGUGCGGUGGUUCAAAG----- -------------.(((((..((((.(((.(((((-(.(....)(((((((((....)))))))))...............)))))).))).))))..)))))...----- ( -29.20, z-score = -3.69, R) >droSec1.super_0 10097775 92 + 21120651 -------------CUGGGCUGUCGUCUACUUGCAU-AUGGCGUCGACAUUGAACUCGUUCAAUGUCAAAAUUAUAAUAAAUUAUGCGUGUGUGCGGCGGUUCAAAG----- -------------.(((((((((((.(((.(((((-(.(....)(((((((((....)))))))))...............)))))).))).)))))))))))...----- ( -34.10, z-score = -5.06, R) >droYak2.chr3R 13323620 87 + 28832112 -------------GCUGUCGGCUGUCUACUUGCAU-AUAGCGUCGACAUUGAACUCGUUCAAUGUCAAAAUUAUAAUAAAUUAUGCGUGUGCGGCUCCAAG---------- -------------.((...((((((.(((..((((-........(((((((((....)))))))))..((((......))))))))))).))))))...))---------- ( -23.10, z-score = -2.08, R) >droEre2.scaffold_4770 10448102 80 - 17746568 --------------------GCUGUCUACUUGCAU-AUGGCGUCGACAUUGAACUCGUUCAAUGUCAAAAUUAUAAUAAAUUAUGCGUGUGCGGCUCCAAG---------- --------------------(((((.(((..((((-(.(....)(((((((((....)))))))))...............)))))))).)))))......---------- ( -22.20, z-score = -2.74, R) >dp4.chr2 20891719 100 - 30794189 CCAAAGCAAAGCGUUUGUCUACUGCUUGUGCAUAUCAUCGACUCGACAUUGAACUCGUUCAAUGUCAAAAUUAUAAUAAAUUAUGCGUGUGUGCUCGGAG----------- ((..((((..((((..(((...(((....))).......)))..(((((((((....)))))))))..................))))...)))).))..----------- ( -24.50, z-score = -1.72, R) >droPer1.super_3 3643660 100 - 7375914 CCAAAGCAAAGCGUUUGUCUACUGCUUGUGCAUAUCAUCGACUCGACAUUGAACUCGUUCAAUGUCAAAAUUAUAAUAAAUUAUGCGUGUGUGCUCGGAG----------- ((..((((..((((..(((...(((....))).......)))..(((((((((....)))))))))..................))))...)))).))..----------- ( -24.50, z-score = -1.72, R) >consensus _____________CUGGGCUGCUGUCUACUUGCAU_AUGGCGUCGACAUUGAACUCGUUCAAUGUCAAAAUUAUAAUAAAUUAUGCGUGUGUGCGUCGAAG__________ .............................((((((....((((.(((((((((....)))))))))..((((......))))))))..))))))................. (-15.11 = -15.01 + -0.10)

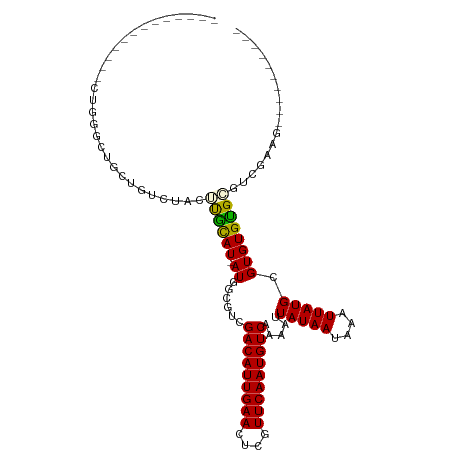
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:36 2011