
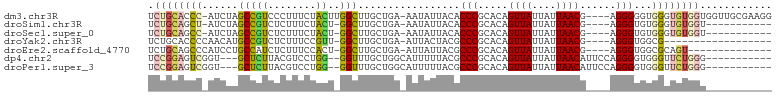
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,895,229 – 10,895,327 |
| Length | 98 |
| Max. P | 0.520447 |

| Location | 10,895,229 – 10,895,327 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 71.45 |
| Shannon entropy | 0.49028 |
| G+C content | 0.50816 |
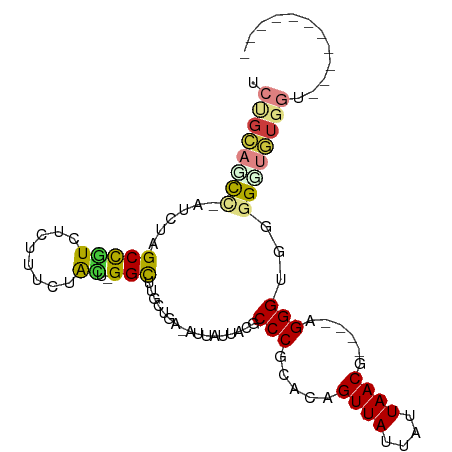
| Mean single sequence MFE | -28.51 |
| Consensus MFE | -11.43 |
| Energy contribution | -11.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10895229 98 + 27905053 UCUGCACCC-AUCUAGCCGUCCCUUUCUACUUGGCUUGCUGA-AAUAUUACACCCGCACAGUUAUUAUUAACG----AGGGGGUGGGUGUGGUGGUUGCGAAGG ..((((.((-(((.(((((............)))))......-......((((((((.(.((((....)))).----..)..))))))))))))).)))).... ( -31.90, z-score = -1.01, R) >droSim1.chr3R 16953632 86 - 27517382 UCUGCAGCU-AUCUAGCCGUCUCUUUCUACU-GGCUUGCUGA-AAUAUUACACCCGCACAGUUAUUAUUAACG----AGGGUGUGGGUGUGGU----------- ....((((.-....((((((........)).-)))).)))).-...(((((((((((((.((((....)))).----...)))))))))))))----------- ( -32.30, z-score = -3.59, R) >droSec1.super_0 10074718 86 - 21120651 UCUGCAGCC-AUCUAGCCGUCUCUUUCUACU-GGCUUGCUGA-AAUAUUACACCCGCACAGUUAUUAUUAACG----AGGGUGUGGGUGUGGU----------- ....((((.-....((((((........)).-)))).)))).-...(((((((((((((.((((....)))).----...)))))))))))))----------- ( -31.60, z-score = -3.14, R) >droYak2.chr3R 13305925 80 - 28832112 UCUGCACCCCAACAUGCCGUCUCUUUCCGUU-GGCUUGCUGA-AUUACUACGCCCGCACAGUUAUUAUUAACG----AGGGUGGCG------------------ ((.(((..(((((...............)))-))..))).))-.......(((((.....((((....)))).----.)))))...------------------ ( -19.96, z-score = -0.68, R) >droEre2.scaffold_4770 10428706 84 + 17746568 UCUGCAGCCCAUCCUGCCAUCUCUUUCCACU-GGCUUGCUGA-AUUAUUACGCCCGCACAGUUAUUAUUAACG----AGGGUGGCGCAGU-------------- .((((.(((((.(..((((...........)-)))..).)).-........((((.....((((....)))).----.))))))))))).-------------- ( -22.20, z-score = -0.66, R) >dp4.chr2 20867704 88 + 30794189 UCCGGAGUCGGU---GCUCUUACGUCCUGG--GGUUUGCUGGCAUUUUUACGCCCGCACAGUUAUUAUUAACAUUCCAGGGGUGGGUUCUGGG----------- .((((((.....---...(((((.((((((--(((.(((.(((........))).)))..((((....)))))))))))))))))))))))).----------- ( -30.80, z-score = -1.77, R) >droPer1.super_3 3620950 88 + 7375914 UCCGGAGUCGGU---GCUCUUACGUCCUGG--GGUUUGCUGGCAUUUUUACGCCCGCACAGUUAUUAUUAACAUUCCAGGGGUGGGUUCUGGG----------- .((((((.....---...(((((.((((((--(((.(((.(((........))).)))..((((....)))))))))))))))))))))))).----------- ( -30.80, z-score = -1.77, R) >consensus UCUGCAGCC_AUCUAGCCGUCUCUUUCUACU_GGCUUGCUGA_AUUAUUACGCCCGCACAGUUAUUAUUAACG____AGGGUGGGGGUGUGGU___________ .((((((((......(((..............))).................(((.....((((....))))......)))...))))))))............ (-11.43 = -11.98 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:34 2011