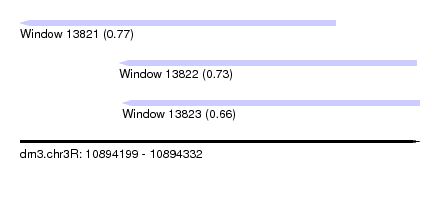
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,894,199 – 10,894,332 |
| Length | 133 |
| Max. P | 0.765972 |

| Location | 10,894,199 – 10,894,304 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Shannon entropy | 0.39416 |
| G+C content | 0.49636 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.04 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10894199 105 - 27905053 ------CCGGAGCCAAAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAGC------GGGCUAAAGCUGAUGCUCAGCUAAUAGUUUGG ------(((((((....))(((((((.((....((.....))..))...)))))))..)))))..(((((.(((((...------.))))).(((((.....)))))....))))). ( -33.40, z-score = -1.55, R) >droPer1.super_3 3620187 83 - 7375914 -----------------------------GAGGAAGAUGAUGAAGGAUAUGCGACUUAUCUGGUUGUAACAUGGCCAAC-----GGGGCCAAAGCUGAUGCUCAGCUAAUAGUUUGG -----------------------------....................(((((((.....)))))))...(((((...-----..))))).(((((.....))))).......... ( -21.50, z-score = -1.04, R) >dp4.chr2 20866932 83 - 30794189 -----------------------------GAGGAAGAUGAUGAAGGAUAUGCGACUUAUCUGGUUGUAACAUGGCCAAC-----GGGGCCAAAGCUGAUGCUCAGCUAAUAGUUUGG -----------------------------....................(((((((.....)))))))...(((((...-----..))))).(((((.....))))).......... ( -21.50, z-score = -1.04, R) >droAna3.scaffold_13340 5971269 110 - 23697760 -------AAGAGCUUGCGCCACCGAACCAGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGCUUCUAACAUGGCCAACGGGCAGGAGCUGAAGCUGAAGCAGAGCUAAUAGUUUUG -------...(((((((.((........((((((((((((((((.....))))..))))))))))))......(((....))).)).))..)))))....((((((.....)))))) ( -34.80, z-score = -1.75, R) >droEre2.scaffold_4770 10427586 111 - 17746568 AAGGAGCCGGAGCCAAAGCAGUUGAAGCUGGGGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAGC------GGGCCAAUGCUGAUGCUCAGCUAAUAGUUUGG .....(((((((((..(((.......)))..)))......((((.....)))).....)))))).(((((.(((((...------.)))))..((((.....)))).....))))). ( -37.40, z-score = -1.59, R) >droYak2.chr3R 13304689 93 + 28832112 ------------------CAGUUGAAGCUGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAGC------GGGCCAAAGCUGAUGCUCAGCUAAUAGUUUGG ------------------((((....)))).(.(((((((((((.....))))..))))))).).(((((.(((((...------.))))).(((((.....)))))....))))). ( -31.10, z-score = -2.42, R) >droSec1.super_0 10073693 105 + 21120651 ------CCGGAGCCAGAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAGC------GGGCUAAAGCUGAUGCUCAGCUAAUAGUUUGG ------.....((((((..(((((((.((....((.....))..))...)))))))..)))))).(((((.(((((...------.))))).(((((.....)))))....))))). ( -37.40, z-score = -2.61, R) >droSim1.chr3R 16952606 105 + 27517382 ------CCGGAGCCAGAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAGC------GGGCCAAAGCUGAUGCUCAGCUAAUAGUUUGG ------.....((((((..(((((((.((....((.....))..))...)))))))..)))))).(((((.(((((...------.))))).(((((.....)))))....))))). ( -39.70, z-score = -3.22, R) >consensus _______CGGAGCCA_AGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAGC______GGGCCAAAGCUGAUGCUCAGCUAAUAGUUUGG .................................(((((((((((.....))))..))))))).....(((.(((((..........))))).(((((.....)))))....)))... (-17.50 = -18.04 + 0.53)

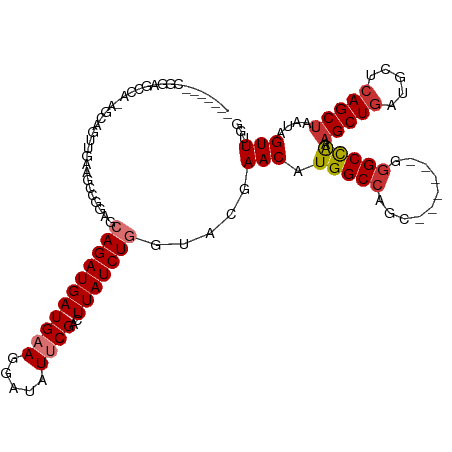
| Location | 10,894,232 – 10,894,331 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.93 |
| Shannon entropy | 0.50792 |
| G+C content | 0.53216 |
| Mean single sequence MFE | -28.85 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
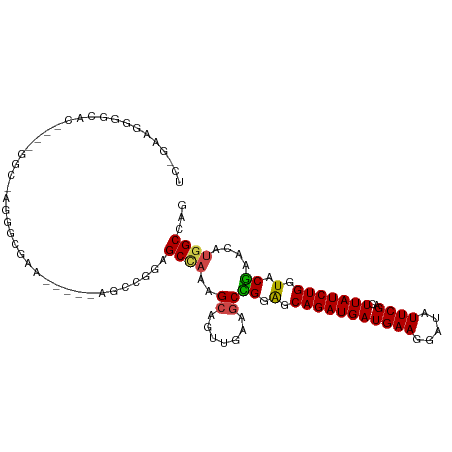
>dm3.chr3R 10894232 99 - 27905053 UCUGAGGGGGCAC----GGCGAGGGCGAA-----AGCCGGAGCCAAAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAG ........(((.(----(((...(((...-----.)))...((....)).......)))).(.(((((((((((.....))))..))))))).).........))).. ( -30.40, z-score = -1.05, R) >droAna3.scaffold_13340 5971308 81 - 23697760 ---------------------------AGGGUGGAAGAAGAGCUUGCGCCACCGAACCAGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGCUUCUAACAUGGCCAA ---------------------------..((((.(((.....))).)))).(((....((((((((((((((((.....))))..))))))))))))....))).... ( -27.30, z-score = -2.62, R) >droEre2.scaffold_4770 10427619 108 - 17746568 UCUGAAGGAAGGCAAAAGGCGAAGCCGGAGAAGGAGCCGGAGCCAAAGCAGUUGAAGCUGGGGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAG ..........(((....(((....((((........)))).)))....((((....)))).(.(((((((((((.....))))..))))))).).........))).. ( -28.70, z-score = -0.61, R) >droYak2.chr3R 13304722 78 + 28832112 ------------------------------UCUGAGGGAAGGCAAAAGCAGUUGAAGCUGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAG ------------------------------..........(((.....((((....)))).(.(((((((((((.....))))..))))))).).........))).. ( -16.90, z-score = -0.31, R) >droSec1.super_0 10073726 99 + 21120651 UCGGAAGGGGCAC----GGCAAGGGCGAA-----AGCCGGAGCCAGAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAG ((....)).....----(((...(((...-----.))).(.((((((..(((((((.((....((.....))..))...)))))))..)))))).).......))).. ( -34.90, z-score = -2.96, R) >droSim1.chr3R 16952639 99 + 27517382 UCGGAAGGGGCAC----GGCAAGGGCGAA-----AGCCGGAGCCAGAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAG ((....)).....----(((...(((...-----.))).(.((((((..(((((((.((....((.....))..))...)))))))..)))))).).......))).. ( -34.90, z-score = -2.96, R) >consensus UC_GAAGGGGCAC____GGC_AGGGCGAA_____AGCCGGAGCCAAAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCAG .........................................((((..((.......))((.(.(((((((((((.....))))..))))))).).))....))))... (-12.69 = -12.50 + -0.19)
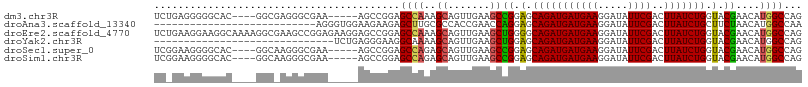

| Location | 10,894,233 – 10,894,332 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 71.58 |
| Shannon entropy | 0.51541 |
| G+C content | 0.53318 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
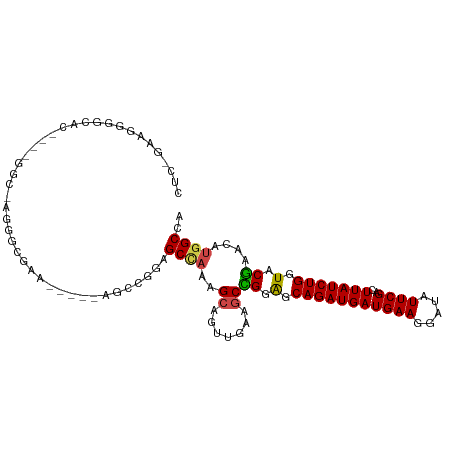
>dm3.chr3R 10894233 99 - 27905053 CUCUGAGGGGGCAC----GGCGAGGGCGAA-----AGCCGGAGCCAAAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCA .........(((.(----(((...(((...-----.)))...((....)).......)))).(.(((((((((((.....))))..))))))).).........))). ( -30.40, z-score = -0.72, R) >droAna3.scaffold_13340 5971309 80 - 23697760 ----------------------------AGGGUGGAAGAAGAGCUUGCGCCACCGAACCAGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGCUUCUAACAUGGCCA ----------------------------..((((.(((.....))).)))).(((....((((((((((((((((.....))))..))))))))))))....)))... ( -27.30, z-score = -2.55, R) >droEre2.scaffold_4770 10427620 108 - 17746568 CUCUGAAGGAAGGCAAAAGGCGAAGCCGGAGAAGGAGCCGGAGCCAAAGCAGUUGAAGCUGGGGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCA ...........(((....(((....((((........)))).)))....((((....)))).(.(((((((((((.....))))..))))))).).........))). ( -28.70, z-score = -0.17, R) >droYak2.chr3R 13304723 78 + 28832112 ------------------------------CUCUGAGGGAAGGCAAAAGCAGUUGAAGCUGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCA ------------------------------...........(((.....((((....)))).(.(((((((((((.....))))..))))))).).........))). ( -16.90, z-score = 0.03, R) >droSec1.super_0 10073727 99 + 21120651 CUCGGAAGGGGCAC----GGCAAGGGCGAA-----AGCCGGAGCCAGAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCA (((....)))....----(((...(((...-----.))).(.((((((..(((((((.((....((.....))..))...)))))))..)))))).).......))). ( -38.00, z-score = -3.52, R) >droSim1.chr3R 16952640 99 + 27517382 CUCGGAAGGGGCAC----GGCAAGGGCGAA-----AGCCGGAGCCAGAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCA (((....)))....----(((...(((...-----.))).(.((((((..(((((((.((....((.....))..))...)))))))..)))))).).......))). ( -38.00, z-score = -3.52, R) >consensus CUC_GAAGGGGCAC____GGC_AGGGCGAA_____AGCCGGAGCCAAAGCAGUUGAAGCCGGAGCAGAUGAUGAAGGAUAUUCGACUUAUCUGGUACGAACAUGGCCA ..........................................((((..((.......))((.(.(((((((((((.....))))..))))))).).))....)))).. (-12.69 = -12.50 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:33 2011