| Sequence ID | dm3.chr3R |
|---|---|
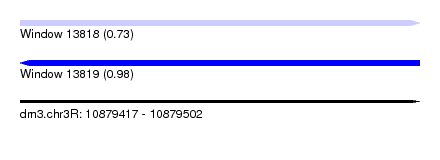
| Location | 10,879,417 – 10,879,502 |
| Length | 85 |
| Max. P | 0.981204 |

| Location | 10,879,417 – 10,879,502 |
|---|---|
| Length | 85 |
| Sequences | 7 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 63.75 |
| Shannon entropy | 0.73896 |
| G+C content | 0.42807 |
| Mean single sequence MFE | -17.93 |
| Consensus MFE | -3.55 |
| Energy contribution | -4.60 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.71 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.729592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
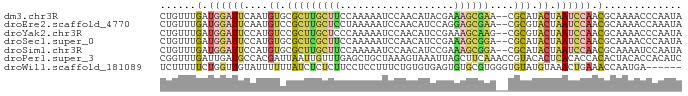
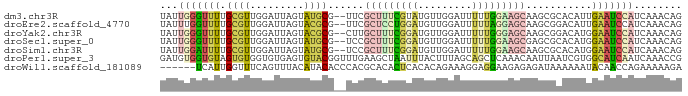
>dm3.chr3R 10879417 85 + 27905053 CUGUUUGAUGGAUUCAAUGUGCGCUUGCUUCCAAAAAUCCAACAUACGAAAGCGAA--CGCAUACUAAUCCAACGCAAAACCCAAUA ...((((.((((((...((((((.((((((.(...............).)))))).--))))))..))))))...))))........ ( -19.96, z-score = -3.34, R) >droEre2.scaffold_4770 10412907 85 + 17746568 CUGUUUGAUGGAUUCAAUGUCCGCUUGCUCCUAAAAAUCCAACAUCCAGGAGCGAA--CGCGUACUAAUCCAACGCAAAACCAAAUA ...((((.((((((....((.(((((((((((...............)))))))).--.))).)).))))))...))))........ ( -21.56, z-score = -3.44, R) >droYak2.chr3R 13290022 85 - 28832112 CUGUUUGAUGGAUUCCAUGUCCGCUUGCUCCCAAAAAUCCAACAUCCGAAAGCAAG--CGCGUACUAAUCCAACGCAAAACCCAAUA ...((((.((((((....(((((((((((..(...............)..))))))--)).).)).))))))...))))........ ( -18.46, z-score = -3.04, R) >droSec1.super_0 10059157 85 - 21120651 CUGUUUGAUGGAUUCCAUGUGCGCUCGCUUCCAAAAAUCCAACAUCCGAAAGCGGA--CGCAUACUAAUCCAACGCAAAACCCAAUA ...((((.((((((...((((((..(((((.(...............).)))))..--))))))..))))))...))))........ ( -22.26, z-score = -3.45, R) >droSim1.chr3R 16936903 85 - 27517382 CUGUUUGAUGGAUUCCAUGUGCGCUUGCUUCCAAAAAUCCAACAUCCGAAAGCGGA--CGCAUACUAAUCCAACGCAAAAUCCAAUA ...((((.((((((...((((((..(((((.(...............).)))))..--))))))..))))))...))))........ ( -19.36, z-score = -2.17, R) >droPer1.super_3 3603755 87 + 7375914 CGGUUUGAUUGAUGCCACGAUUAAUUGUUUGAGCUGCUAAAGUAAAUUAGCUUCAAACCGUACACUCACACCACACUACACCACAUC .(((.(((.(((((............(((((((..(((((......))))))))))))))).)).))).)))............... ( -15.40, z-score = -1.06, R) >droWil1.scaffold_181089 8774481 81 + 12369635 UCUUUUUCUGGUUGUAUUUUUUAUCUCUCUUCCUCCUUUCUGUGUGAGUGUGCGUGGGUGUAUGUAAACUGAAACCAAUGA------ ........(((((.((.(((.((((.(((..(((((.......).))).).....))).).))).))).)).)))))....------ ( -8.50, z-score = 0.80, R) >consensus CUGUUUGAUGGAUUCCAUGUGCGCUUGCUUCCAAAAAUCCAACAUCCGAAAGCGAA__CGCAUACUAAUCCAACGCAAAACCCAAUA ......(.((((((..((((....((((((...................))))))....))))...)))))).)............. ( -3.55 = -4.60 + 1.05)
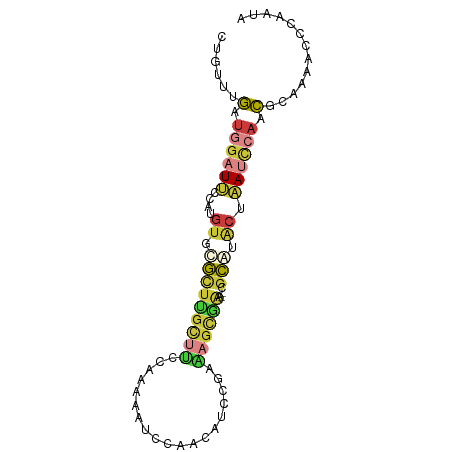
| Location | 10,879,417 – 10,879,502 |
|---|---|
| Length | 85 |
| Sequences | 7 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 63.75 |
| Shannon entropy | 0.73896 |
| G+C content | 0.42807 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.99 |
| Covariance contribution | 0.66 |
| Combinations/Pair | 1.80 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
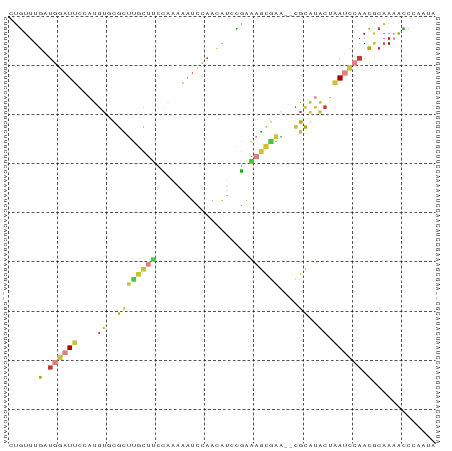
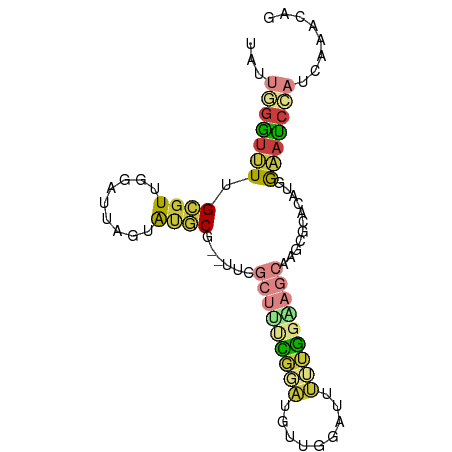
>dm3.chr3R 10879417 85 - 27905053 UAUUGGGUUUUGCGUUGGAUUAGUAUGCG--UUCGCUUUCGUAUGUUGGAUUUUUGGAAGCAAGCGCACAUUGAAUCCAUCAAACAG ........((((...(((((((((.((((--((.(((((((.............))))))).)))))).)))).))))).))))... ( -23.92, z-score = -1.97, R) >droEre2.scaffold_4770 10412907 85 - 17746568 UAUUUGGUUUUGCGUUGGAUUAGUACGCG--UUCGCUCCUGGAUGUUGGAUUUUUAGGAGCAAGCGGACAUUGAAUCCAUCAAACAG ........((((...((((((.((.(((.--...(((((((((.........)))))))))..))).))....)))))).))))... ( -24.10, z-score = -1.83, R) >droYak2.chr3R 13290022 85 + 28832112 UAUUGGGUUUUGCGUUGGAUUAGUACGCG--CUUGCUUUCGGAUGUUGGAUUUUUGGGAGCAAGCGGACAUGGAAUCCAUCAAACAG ...((((((((((((.........))))(--((((((..((((.........))))..)))))))......))))))))........ ( -28.60, z-score = -3.36, R) >droSec1.super_0 10059157 85 + 21120651 UAUUGGGUUUUGCGUUGGAUUAGUAUGCG--UCCGCUUUCGGAUGUUGGAUUUUUGGAAGCGAGCGCACAUGGAAUCCAUCAAACAG ........((((...((((((.((.((((--..((((((((((.........))))))))))..)))).))..)))))).))))... ( -28.40, z-score = -2.66, R) >droSim1.chr3R 16936903 85 + 27517382 UAUUGGAUUUUGCGUUGGAUUAGUAUGCG--UCCGCUUUCGGAUGUUGGAUUUUUGGAAGCAAGCGCACAUGGAAUCCAUCAAACAG ...(((((((((((((((((........)--)))(((((((((.........))))))))).))))))....)))))))........ ( -26.10, z-score = -2.23, R) >droPer1.super_3 3603755 87 - 7375914 GAUGUGGUGUAGUGUGGUGUGAGUGUACGGUUUGAAGCUAAUUUACUUUAGCAGCUCAAACAAUUAAUCGUGGCAUCAAUCAAACCG .....(((......((((((.((......((((((.(((((......)))))...)))))).......).).)))))).....))). ( -20.22, z-score = -0.44, R) >droWil1.scaffold_181089 8774481 81 - 12369635 ------UCAUUGGUUUCAGUUUACAUACACCCACGCACACUCACACAGAAAGGAGGAAGAGAGAUAAAAAAUACAACCAGAAAAAGA ------...((((((...((......))...........(((.(.......))))...................))))))....... ( -5.90, z-score = 0.65, R) >consensus UAUUGGGUUUUGCGUUGGAUUAGUAUGCG__UUCGCUUUCGGAUGUUGGAUUUUUGGAAGCAAGCGCACAUGGAAUCCAUCAAACAG ...(((((((.((((.........))))......(((((((((.........)))))))))...........)))))))........ ( -9.32 = -9.99 + 0.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:30 2011