| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,874,378 – 10,874,503 |
| Length | 125 |
| Max. P | 0.737406 |

| Location | 10,874,378 – 10,874,503 |
|---|---|
| Length | 125 |
| Sequences | 6 |
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 80.10 |
| Shannon entropy | 0.35973 |
| G+C content | 0.45316 |
| Mean single sequence MFE | -34.95 |
| Consensus MFE | -21.34 |
| Energy contribution | -22.35 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.637994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
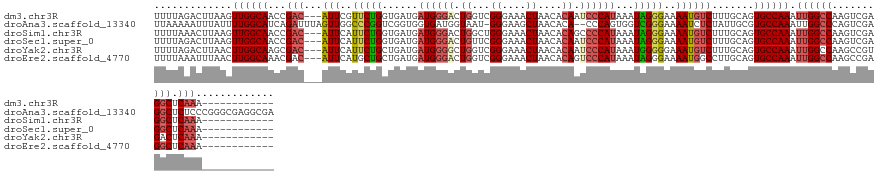
>dm3.chr3R 10874378 125 + 27905053 ------------UUUGAGCCUCGACUUGGCCAAUUUGGCACUGCAAAGACAUUUUCCCUAUUUAUGGGAUUGUGUUAGUUUCCCGACCAGUCCCAUCAUCACCAGAACGAAU---GUCGGUUGCCAACUUAAGUCUAAAA ------------..........(((((((((.....)))...((((.(((((((((.......(((((((((.((..(....)..)))))))))))........))..))))---)))..)))).....))))))..... ( -35.36, z-score = -2.65, R) >droAna3.scaffold_13340 5952347 137 + 23697760 UCGCCUCGCCCGGGAGAGCCUCGACUGGGCCAAUUUGGCACGCAAUAGAGAUUUUCCCGACCACUGGG--UGUGUUAGCUUCCC-AUUACCAUCACCACCGACCGGGCCAACUAAAUCUGAUGCCAAAAUAAAUUUUUAA .......((((((..(.....)..))))))...((((((((......)((((((.((((.....((((--.((....))..)))-)......((......)).))))......))))))..)))))))............ ( -33.50, z-score = 0.36, R) >droSim1.chr3R 16931909 125 - 27517382 ------------UUUGAGCCUCGACUUGGCCAAUUUGGCACUGCAAAGACAUUUUCCCUAUUUAUGGGGCUGUGUUAGUUUCCCAGCCAGUCCCAUCAUCACCAGAAUGAAU---GUCGGUUGCCAACUUAAGUUUAAAA ------------.(((.(((.......)))))).((((((..((...(((((((...((....(((((((((.(((........)))))))))))).......))...))))---))).))))))))............. ( -35.70, z-score = -2.00, R) >droSec1.super_0 10054165 125 - 21120651 ------------UUUGAGCCUCGACUUGGCCAAUUUGGCACUGCAAAGACAUUUUCCCUAUUUAUGGGAUUGUGUUAGUUUCCCGAACAGUCCCAUCAUCACCAGAAUGAAU---GUCGGUUGCCAACUUAAGUCUAAAA ------------..........(((((((((.....)))...((((.(((((((...((....((((((((((.............)))))))))).......))...))))---)))..)))).....))))))..... ( -34.32, z-score = -2.28, R) >droYak2.chr3R 13284922 125 - 28832112 ------------UUUGAGUCACGGCUUGGCCAAUUUGGCACUGCAAAGACAUUUCCCCCAUUUAUGGGAUUGUGUUAGUUUCCCGACCAGCCCCAUCAUCAGCAGAAUGAAU---GUCGCUUGCCAAGUUAAGUCUAAAA ------------..........((((((((....((((((..((...(((((((.........(((((..((.((..(....)..))))..)))))..((....))..))))---))))).))))))))))))))..... ( -31.70, z-score = -0.83, R) >droEre2.scaffold_4770 10407868 125 + 17746568 ------------UUUGAGCCUCGGCUUGGCCAAUUUGGCACUGCAAGGCCAUUUUCCCUAUUUAUGGGACUGUGUUAGUUUCCCGACCAGUCCCAUCAUCAGCAGCAUGAAU---GUCGUUUGCCAAGUUAAAUUUAAAA ------------.(..(((....)))..)..(((((((((((((.(((........)))....(((((((((.((..(....)..))))))))))).....)))).(((...---..))).))))))))).......... ( -39.10, z-score = -2.73, R) >consensus ____________UUUGAGCCUCGACUUGGCCAAUUUGGCACUGCAAAGACAUUUUCCCUAUUUAUGGGAUUGUGUUAGUUUCCCGACCAGUCCCAUCAUCACCAGAAUGAAU___GUCGGUUGCCAACUUAAGUCUAAAA .............(((.(((.......)))))).(((((((((......((((((........(((((((((.(((........)))))))))))).......))))))........))).))))))............. (-21.34 = -22.35 + 1.01)


| Location | 10,874,378 – 10,874,503 |
|---|---|
| Length | 125 |
| Sequences | 6 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 80.10 |
| Shannon entropy | 0.35973 |
| G+C content | 0.45316 |
| Mean single sequence MFE | -40.27 |
| Consensus MFE | -23.22 |
| Energy contribution | -24.42 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.737406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10874378 125 - 27905053 UUUUAGACUUAAGUUGGCAACCGAC---AUUCGUUCUGGUGAUGAUGGGACUGGUCGGGAAACUAACACAAUCCCAUAAAUAGGGAAAAUGUCUUUGCAGUGCCAAAUUGGCCAAGUCGAGGCUCAAA------------ .....(((((......((((..(((---(((..(((((......((((((.((((..(....)..)).)).))))))...)))))..)))))).)))).(.(((.....)))))))))..........------------ ( -39.60, z-score = -2.37, R) >droAna3.scaffold_13340 5952347 137 - 23697760 UUAAAAAUUUAUUUUGGCAUCAGAUUUAGUUGGCCCGGUCGGUGGUGAUGGUAAU-GGGAAGCUAACACA--CCCAGUGGUCGGGAAAAUCUCUAUUGCGUGCCAAAUUGGCCCAGUCGAGGCUCUCCCGGGCGAGGCGA ............((((((((..(((..((((..((((..((.(((((.((....(-((....))).))))--.))).))..))))..)).))..)))..))))))))...(((..(((..((.....)).)))..))).. ( -42.80, z-score = 0.25, R) >droSim1.chr3R 16931909 125 + 27517382 UUUUAAACUUAAGUUGGCAACCGAC---AUUCAUUCUGGUGAUGAUGGGACUGGCUGGGAAACUAACACAGCCCCAUAAAUAGGGAAAAUGUCUUUGCAGUGCCAAAUUGGCCAAGUCGAGGCUCAAA------------ .......((..(.(((((....(((---(((..(((((......(((((.((((.(((....))).).))).)))))...)))))..))))))....((((.....))))))))).)..)).......------------ ( -37.80, z-score = -1.66, R) >droSec1.super_0 10054165 125 + 21120651 UUUUAGACUUAAGUUGGCAACCGAC---AUUCAUUCUGGUGAUGAUGGGACUGUUCGGGAAACUAACACAAUCCCAUAAAUAGGGAAAAUGUCUUUGCAGUGCCAAAUUGGCCAAGUCGAGGCUCAAA------------ .....(((((......((((..(((---(((..(((((......((((((.(((..((....))...))).))))))...)))))..)))))).)))).(.(((.....)))))))))..........------------ ( -36.20, z-score = -1.75, R) >droYak2.chr3R 13284922 125 + 28832112 UUUUAGACUUAACUUGGCAAGCGAC---AUUCAUUCUGCUGAUGAUGGGGCUGGUCGGGAAACUAACACAAUCCCAUAAAUGGGGGAAAUGUCUUUGCAGUGCCAAAUUGGCCAAGCCGUGACUCAAA------------ .....((.(((.(((((((((.(((---(((...(((.(((...(((((..((((..(....)..)).))..)))))...))).))))))))))))))...(((.....)))))))...))).))...------------ ( -40.20, z-score = -2.16, R) >droEre2.scaffold_4770 10407868 125 - 17746568 UUUUAAAUUUAACUUGGCAAACGAC---AUUCAUGCUGCUGAUGAUGGGACUGGUCGGGAAACUAACACAGUCCCAUAAAUAGGGAAAAUGGCCUUGCAGUGCCAAAUUGGCCAAGCCGAGGCUCAAA------------ ............((((((.......---......(((((.....(((((((((((..(....)..)).)))))))))....((((.......)))))))))(((.....)))...)))))).......------------ ( -45.00, z-score = -3.86, R) >consensus UUUUAAACUUAAGUUGGCAACCGAC___AUUCAUUCUGGUGAUGAUGGGACUGGUCGGGAAACUAACACAAUCCCAUAAAUAGGGAAAAUGUCUUUGCAGUGCCAAAUUGGCCAAGUCGAGGCUCAAA____________ .............((((((..(((....(((..(((((......((((((.((((.((....)).)).)).))))))...)))))..)))....)))...)))))).((((((.......))).)))............. (-23.22 = -24.42 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:28 2011