
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,836,958 – 10,837,050 |
| Length | 92 |
| Max. P | 0.508596 |

| Location | 10,836,958 – 10,837,050 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.15 |
| Shannon entropy | 0.58993 |
| G+C content | 0.49440 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.03 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10836958 92 + 27905053 ----------------------------ACAGAAAAAGAGGAGACUACGGACUGCGGACUGCGGCACAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCGCGAAUUGUGCAC ----------------------------..........((....))..(..((((.....))))..)....((((((((..((.....))..)))))))).....((((.....)))).. ( -23.00, z-score = -0.28, R) >droSim1.chr3R 16897717 92 - 27517382 ----------------------------ACAGAAAAAGAGGAGACUACGGACUGCGGACUGCGGCACAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCGCGAAUUGUGCAC ----------------------------..........((....))..(..((((.....))))..)....((((((((..((.....))..)))))))).....((((.....)))).. ( -23.00, z-score = -0.28, R) >droSec1.super_0 10020654 92 - 21120651 ----------------------------ACAGAAAAAGAGGAGACUACGGACUGCGGACUGCGGCACAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCGCGAAUUGUGCAC ----------------------------..........((....))..(..((((.....))))..)....((((((((..((.....))..)))))))).....((((.....)))).. ( -23.00, z-score = -0.28, R) >droYak2.chr3R 13245442 85 - 28832112 ----------------------------ACAGAAAAAGAGGAGA-------CUGCGGACUGCGGCACAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCGCGAAUUGUGCAC ----------------------------.(((.....(((....-------)).)...)))..((((((((((((((((..((.....))..))))))((.....))))))).))))).. ( -21.80, z-score = -0.23, R) >droEre2.scaffold_4770 10373967 92 + 17746568 ----------------------------ACAGAAAAAGAGGAGACUGCAGACUGCGGACUGCGGCACAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCGCGAAUUGUGCAC ----------------------------..........((....))((((........)))).((((((((((((((((..((.....))..))))))((.....))))))).))))).. ( -25.70, z-score = -0.64, R) >droAna3.scaffold_13340 5918100 89 + 23697760 -----------------------------UUGGGAGGCAGGGAGC--CGGACUCCGGACUGCGGCACAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCACGAAUUGUGCGA -----------------------------.......((((....(--(((...)))).)))).((((((((((((((((..((.....))..))))))((.....))))))).))))).. ( -29.60, z-score = -1.29, R) >dp4.chr2 20804027 120 + 30794189 GUGGAGAUGGAGAUGAUGGCGAGGCGGAGACGGGUGGGGUGAACGCAUGGGCGAAGGACUGCGGCACAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCGCGAAUUGUGCAC (((..((((((((..(((((....((....)).(..(..(...(((....)))..)..)..).)).)).)..)))))))).((((((.((((((....)))).....)).)))))).))) ( -36.10, z-score = -1.43, R) >droPer1.super_3 3556020 120 + 7375914 GUGGAGAUGGAGAUGAUGGCAAGGCGGAGACGGGUGGGGUGAACGCAUGGGCGAAGGACUGCGGCACAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCGCGAAUCGUGCAC ((.((..((..(((((..(((((.((....))......(((..((((.(..(....).))))).))).)))))..)))))..)).)).))((((....))))...(((((...))))).. ( -35.70, z-score = -1.10, R) >droWil1.scaffold_181089 8737342 79 + 12369635 -------------------------------------AAAAGCGGCAAUGGCAAAGGA----GAAGCAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCGCGAAUUGUGCAC -------------------------------------((((((.((((..((......----...))..))))((((((..((.....))..)))))))))))).((((.....)))).. ( -19.40, z-score = -0.09, R) >droVir3.scaffold_13047 242959 90 - 19223366 -------------------------GUGGUGUUGGGGGCAAGC--UCAAAGCUAUCGAC---UGCGCAUUUGUUUUCAUCGGCAAUUGGCAAGCGAAAGCUUUUAGCGGAAAUGGUGCAC -------------------------...((((.........((--(.((((((.(((..---(((.((.(((((......))))).)))))..))).)))))).))).........)))) ( -27.07, z-score = -0.72, R) >droGri2.scaffold_14906 2182043 92 - 14172833 -------------------------GCAGUGCUGGGGGGAACACGUCAAUCCCAUGGAC---UGCGCAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCAGAAAUUGUGCAC -------------------------(((((.((((((.((.....))..))))).).))---)))((((..((((((((..((.....))..))))))))..............)))).. ( -28.59, z-score = -1.08, R) >droMoj3.scaffold_6540 14707817 90 + 34148556 -------------------------ACGGUAUUAGGGGCUGGC--GCAAGGCAAUAGAC---GGCGCAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAACGGGAAUUGUGCAC -------------------------.((((.......))))((--((((..(......(---(........((((((((..((.....))..))))))))......)).)..)))))).. ( -22.24, z-score = -0.44, R) >consensus ____________________________ACAGAAAGGGAGGAGACUACGGACUACGGACUGCGGCACAUUUGUUUUCAUCGGCAAUUGGCAAGUGAAAGCUUUUAGCGCGAAUUGUGCAC ...............................................................((((((((((((((((..((.....))..)))))))).........)))).)))).. (-14.14 = -14.03 + -0.11)
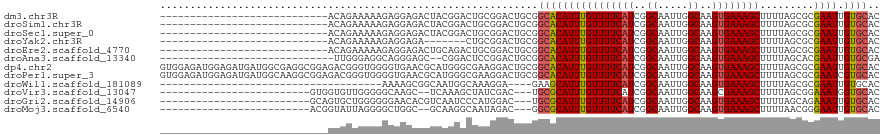

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:24 2011