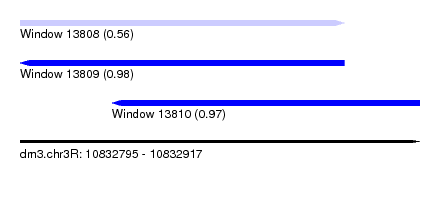
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,832,795 – 10,832,917 |
| Length | 122 |
| Max. P | 0.978343 |

| Location | 10,832,795 – 10,832,894 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Shannon entropy | 0.51337 |
| G+C content | 0.40046 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -13.02 |
| Energy contribution | -13.23 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
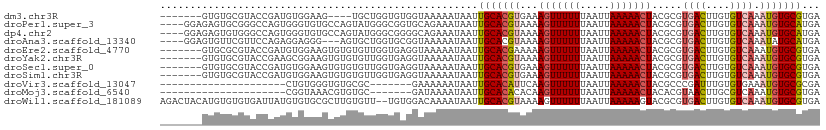
>dm3.chr3R 10832795 99 + 27905053 -------GUGUGCGUACCGAUGUGGAAG----UGCUGGUGUGGUAAAAAUAAUUGCACGUGAAAGUUUUUUAAUUAAAAACUACGCGUGACUUGUGUCAAAUGUGCGUGA -------....((((((..(((((...(----(((...(((.......)))...)))).....(((((((.....))))))).)))))(((....)))....)))))).. ( -24.60, z-score = -0.97, R) >droPer1.super_3 3551370 106 + 7375914 ----GGAGAGUGCGGGCCAGUGGGUGUGCCAGUAUGGGCGGUGCAGAAAUAAUUGCACGUAAAAGUUUUUUAAUUAAAAACUACGCGUGACUUGUGUCAAAUGUGCAUGA ----.....(..(.(.(((...((....))....))).).)..).........(((((((...(((((((.....))))))).....((((....)))).)))))))... ( -30.40, z-score = -1.01, R) >dp4.chr2 20799381 106 + 30794189 ----GGAGAGUGUGGGCCAGUGGGUGUGCCAGUAUGGGCGGGGCAGAAAUAAUUGCACGUAAAAGUUUUUUAAUUAAAAACUACGCGUGACUUGUGUCAAAUGUGCAUGA ----.....((.(.(.(((...((....))....))).).).)).........(((((((...(((((((.....))))))).....((((....)))).)))))))... ( -28.00, z-score = -0.59, R) >droAna3.scaffold_13340 5914396 103 + 23697760 ----GGAGUGUUCGUUCCAGAGGAGGG---AGUGCUGGUGCGGUAAAAAUAAUUGCACGUAAAAGUUUUUUAAUUAAAAACUACGCGUGACUUGUGUCAAAUAUGCAUGA ----..((..(((.((((...)))).)---))..)).(((((((.......))))))).....(((((((.....)))))))..(((((((....))))....))).... ( -27.00, z-score = -1.84, R) >droEre2.scaffold_4770 10369789 103 + 17746568 -------GUGCGCGUACCGAUGUGGAAGUGUGUGUUGGUGAGGUAAAAAUAAUUGCACGAAAAAGUUUUUUAAUUAAAAACUACGCGUGACUUGUGUCAAAUGUGCGUGA -------...(((((((..(((((...((((.((((...........))))...)))).....(((((((.....))))))).)))))(((....)))....))))))). ( -24.10, z-score = -0.55, R) >droYak2.chr3R 13241189 103 - 28832112 -------GUGUGCGUACCGAAGCGGAAGUGUGUGUUGGUGAGGUAAAAAUAAUUGCACGUAAAAGUUUUUUAAUUAAAAACUACGCGUGACUUGUGUCAAAUGUGCGUGA -------...(((.((((((.((....)).....))))))..)))..........(((((...(((((((.....)))))))(((..((((....))))..)))))))). ( -26.20, z-score = -1.27, R) >droSec1.super_0 10016476 103 - 21120651 -------GUGUGCGUACCGAUGUGGAAGUGUGUGUUGGUGAGGUAAAAAUAAUUGCACGUGAAAGUUUUUUAAUUAAAAACUACGCGUGACUUGUGUCAAAUGUGCGUGA -------...(((.((((((..(........)..))))))..))).........(((((((..(((((((.....))))))).))).((((....))))...)))).... ( -24.70, z-score = -1.04, R) >droSim1.chr3R 16893485 103 - 27517382 -------GUGUGCGUACCGAUGUGGAAGUGUGUGUUGGUGAGGUAAAAAUAAUUGCACGUGAAAGUUUUUUAAUUAAAAACUACGCGUGACUUGUGUCAAAUGUGCGUGA -------...(((.((((((..(........)..))))))..))).........(((((((..(((((((.....))))))).))).((((....))))...)))).... ( -24.70, z-score = -1.04, R) >droVir3.scaffold_13047 239017 82 - 19223366 ---------------------CUGUGGGUGUGCGC-------GAAAAAAUAAUUGCACAUUCAAGUUUUUUAAUUAAAAACUACGCCCGAUUUGUGUGAAAUGUGCGCGA ---------------------...((((((((.((-------((........)))).).....(((((((.....))))))))))))))..((((((.......)))))) ( -20.50, z-score = -1.20, R) >droMoj3.scaffold_6540 14703784 82 + 34148556 ---------------------CGGUAAACGUGUGC-------GAUAAAAUAAUUGCACACACAAGUUUUUUAAUUAAAAACUACACGUAACUUGCGUCAAAUGUGCGUGA ---------------------........((((((-------(((......)))))))))...(((((((.....))))))).((((((..((......))..)))))). ( -23.00, z-score = -2.35, R) >droWil1.scaffold_181089 8733434 108 + 12369635 AGACUACAUGUGUGUGAUUAUGUGUGCGCUUGUGUU--UGUGGACAAAAUAAUUGCACGUAAAAGUUUUUUAAUUAAAAAGUACGCGUGACUUGUGUCAAAUGUGCGUGA (((((...((((((..(((((.(((.(((.......--.))).)))..)))))..))))))..)))))............(((((..((((....))))..))))).... ( -29.00, z-score = -1.46, R) >consensus _______GUGUGCGUACCGAUGUGGAAGUGUGUGUUGGUGAGGUAAAAAUAAUUGCACGUAAAAGUUUUUUAAUUAAAAACUACGCGUGACUUGUGUCAAAUGUGCGUGA .....................................................(((((((...(((((((.....))))))).....((((....)))).)))))))... (-13.02 = -13.23 + 0.21)
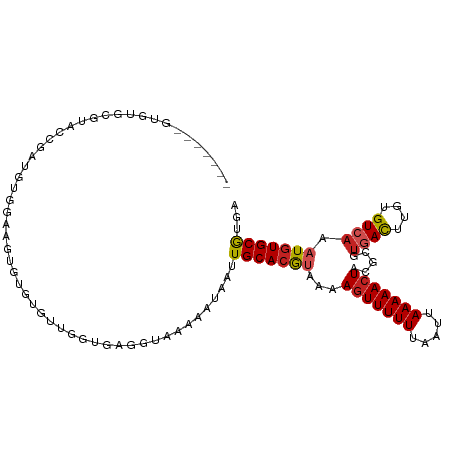
| Location | 10,832,795 – 10,832,894 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.09 |
| Shannon entropy | 0.51337 |
| G+C content | 0.40046 |
| Mean single sequence MFE | -16.02 |
| Consensus MFE | -11.15 |
| Energy contribution | -11.18 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10832795 99 - 27905053 UCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUCACGUGCAAUUAUUUUUACCACACCAGCA----CUUCCACAUCGGUACGCACAC------- ...........((((....)))).(((((((((((.....))))))......((((...................)))----)...........)))))....------- ( -15.61, z-score = -2.19, R) >droPer1.super_3 3551370 106 - 7375914 UCAUGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUCUGCACCGCCCAUACUGGCACACCCACUGGCCCGCACUCUCC---- ...(((.....((((....)))).(((.(((((((.....))))))).....(((((........)))))))).......(((.((.....))))).)))......---- ( -20.90, z-score = -1.82, R) >dp4.chr2 20799381 106 - 30794189 UCAUGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUCUGCCCCGCCCAUACUGGCACACCCACUGGCCCACACUCUCC---- ...(((((...((((....))))...(((((((((.....)))))))...)))))))..............(((......))).......................---- ( -17.50, z-score = -1.09, R) >droAna3.scaffold_13340 5914396 103 - 23697760 UCAUGCAUAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUUUACCGCACCAGCACU---CCCUCCUCUGGAACGAACACUCC---- ((.(((.....((((....))))((((((((((((.....)))))))...)))))..............)))((((....---.......))))...)).......---- ( -17.10, z-score = -2.04, R) >droEre2.scaffold_4770 10369789 103 - 17746568 UCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUUCGUGCAAUUAUUUUUACCUCACCAACACACACUUCCACAUCGGUACGCGCAC------- ....((.....((((....)))).(((((((((((.....))))))......(((.....................)))...............)))))))..------- ( -15.00, z-score = -1.69, R) >droYak2.chr3R 13241189 103 + 28832112 UCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUUUACCUCACCAACACACACUUCCGCUUCGGUACGCACAC------- ....((.....((((....))))((((((((((((.....)))))))...)))))................................((....))..))....------- ( -15.40, z-score = -1.73, R) >droSec1.super_0 10016476 103 + 21120651 UCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUCACGUGCAAUUAUUUUUACCUCACCAACACACACUUCCACAUCGGUACGCACAC------- ....((.....((((....))))((((((((((((.....)))))))...)))))..........((((.......................)))).))....------- ( -14.20, z-score = -2.01, R) >droSim1.chr3R 16893485 103 + 27517382 UCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUCACGUGCAAUUAUUUUUACCUCACCAACACACACUUCCACAUCGGUACGCACAC------- ....((.....((((....))))((((((((((((.....)))))))...)))))..........((((.......................)))).))....------- ( -14.20, z-score = -2.01, R) >droVir3.scaffold_13047 239017 82 + 19223366 UCGCGCACAUUUCACACAAAUCGGGCGUAGUUUUUAAUUAAAAAACUUGAAUGUGCAAUUAUUUUUUC-------GCGCACACCCACAG--------------------- ......................(((.(((((((((.....))))))).....((((............-------)))))).)))....--------------------- ( -13.50, z-score = -0.71, R) >droMoj3.scaffold_6540 14703784 82 - 34148556 UCACGCACAUUUGACGCAAGUUACGUGUAGUUUUUAAUUAAAAAACUUGUGUGUGCAAUUAUUUUAUC-------GCACACGUUUACCG--------------------- ....((((((.((((....)))).))))(((((((.....))))))).))((((((.((......)).-------))))))........--------------------- ( -20.40, z-score = -2.00, R) >droWil1.scaffold_181089 8733434 108 - 12369635 UCACGCACAUUUGACACAAGUCACGCGUACUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUUGUCCACA--AACACAAGCGCACACAUAAUCACACACAUGUAGUCU ..((((.....((((....)))).))))........................((((..((..((((....))--))...))..))))....................... ( -12.40, z-score = 0.14, R) >consensus UCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUUUACCUCACCAACACACACUUCCACAUCGGUACGCACAC_______ ....((((...((((....)))).....(((((((.....))))))).....))))...................................................... (-11.15 = -11.18 + 0.04)
| Location | 10,832,823 – 10,832,917 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.10 |
| Shannon entropy | 0.35153 |
| G+C content | 0.31800 |
| Mean single sequence MFE | -13.23 |
| Consensus MFE | -11.15 |
| Energy contribution | -11.18 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10832823 94 - 27905053 ---------UUUUCA--UCUUUUUUUGCCACCUUUCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUCACGUGCAAUUAUUUUUACCAC ---------......--.....................((((...((((....)))).(.(.(((((((.....)))))))..).)))))............... ( -13.10, z-score = -2.72, R) >droPer1.super_3 3551405 105 - 7375914 CGUAUGUAUUUUUCAUUUUUUUUUUUGCCACCUUUCAUGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUCUGCACC .(((((((............................((((.....((((....)))).))))(((((((.....)))))))..)))))))............... ( -14.70, z-score = -1.15, R) >dp4.chr2 20799416 102 - 30794189 CGUAUGUAUUUUUCA---UUUUUUUUGCCACCUUUCAUGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUCUGCCCC .(((((((.......---..................((((.....((((....)))).))))(((((((.....)))))))..)))))))............... ( -14.70, z-score = -1.48, R) >droAna3.scaffold_13340 5914428 92 - 23697760 ---------UUUUCA----UUUUUUUGCCACCUUUCAUGCAUAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUUUACCGC ---------......----..................(((.....((((....)))).(((((((((((.....)))))))...))))))).............. ( -12.20, z-score = -1.34, R) >droEre2.scaffold_4770 10369821 94 - 17746568 ---------UUUUCA--AUCUUUUUUGCCAGCUUUCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUUCGUGCAAUUAUUUUUACCUC ---------......--.......(((((.......((((.....((((....)))).))))(((((((.....))))))).....).))))............. ( -12.60, z-score = -1.70, R) >droYak2.chr3R 13241221 92 + 28832112 ---------UUUUCA----UUUUUUUGCCACCUUUCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUUUACCUC ---------......----...................((((...((((....))))...(((((((((.....)))))))...))))))............... ( -12.70, z-score = -2.44, R) >droSec1.super_0 10016508 92 + 21120651 ---------UUUUCA----UUUUUUUGCCACCUUUCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUCACGUGCAAUUAUUUUUACCUC ---------......----...................((((...((((....)))).(.(.(((((((.....)))))))..).)))))............... ( -13.10, z-score = -2.71, R) >droSim1.chr3R 16893517 92 + 27517382 ---------UUUUCA----UUUUUUUGCCACCUUUCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUCACGUGCAAUUAUUUUUACCUC ---------......----...................((((...((((....)))).(.(.(((((((.....)))))))..).)))))............... ( -13.10, z-score = -2.71, R) >droVir3.scaffold_13047 239033 80 + 19223366 --------------------UAUUUUGUCACCUUUCGCGCACAUUUCACACAAAUCGGGCGUAGUUUUUAAUUAAAAAACUUGAAUGUGCAAUUAUUUUU----- --------------------....(((.(((..(((((((..((((.....))))...))))(((((((.....))))))).))).))))))........----- ( -12.40, z-score = -0.92, R) >droMoj3.scaffold_6540 14703800 84 - 34148556 -----------UUCA-----CAUUUCGUUACCUUUCACGCACAUUUGACGCAAGUUACGUGUAGUUUUUAAUUAAAAAACUUGUGUGUGCAAUUAUUUUA----- -----------....-----...............((((((((..((((....)))).....(((((((.....)))))))))))))))...........----- ( -17.70, z-score = -2.06, R) >droWil1.scaffold_181089 8733471 91 - 12369635 ----------AUUCA----UUUUCUCUUAGUUUUUCACGCACAUUUGACACAAGUCACGCGUACUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUUGUCCAC ----------.....----...................((((...((((....))))...(((.(((((.....)))))....)))))))............... ( -9.20, z-score = -0.78, R) >consensus _________UUUUCA____UUUUUUUGCCACCUUUCACGCACAUUUGACACAAGUCACGCGUAGUUUUUAAUUAAAAAACUUUUACGUGCAAUUAUUUUUACCUC ......................................((((...((((....)))).....(((((((.....))))))).....))))............... (-11.15 = -11.18 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:23 2011