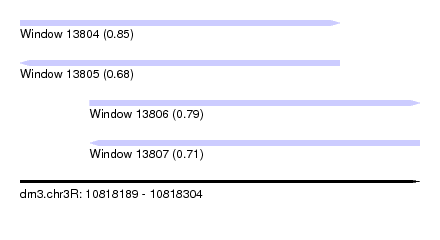
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,818,189 – 10,818,304 |
| Length | 115 |
| Max. P | 0.854441 |

| Location | 10,818,189 – 10,818,281 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.77 |
| Shannon entropy | 0.48397 |
| G+C content | 0.55373 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -15.29 |
| Energy contribution | -14.81 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
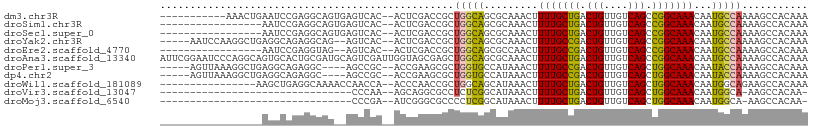
>dm3.chr3R 10818189 92 + 27905053 -------GCCGCCUCGUCUG----GGCCAUGUUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCAGCAAAAGUUUGCGCUGCCAGC--------GGUCGAGUGUG -------..(((((((.(((----((((((....)))))).((((((.(((..((..((((((....))))))....))..))).)))))))--------)).)))).))) ( -40.60, z-score = -2.55, R) >droSim1.chr3R 16878920 92 - 27517382 -------GCCGCCUCGUCUG----GGCCAUGUUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCAGCAAAAGUUUGCGCUGCCAGC--------GGUCGAGUGUG -------..(((((((.(((----((((((....)))))).((((((.(((..((..((((((....))))))....))..))).)))))))--------)).)))).))) ( -40.60, z-score = -2.55, R) >droSec1.super_0 10002033 92 - 21120651 -------GCCGCCUCGUCUG----GGCCAUGUUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCAGCAAAAGUUUGCGCUGCCAGC--------GGUCGAGUGUG -------..(((((((.(((----((((((....)))))).((((((.(((..((..((((((....))))))....))..))).)))))))--------)).)))).))) ( -40.60, z-score = -2.55, R) >droYak2.chr3R 13225515 92 - 28832112 -------GCCGCCUCGUCUG----GGCCAUGUUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCGGCAAAAGUUUGCGCUGCCAGC--------GGUCGAGUGUG -------..(((((((.(((----((((((....)))))).((((((.(((((((((((((....))))))))))......))).)))))))--------)).)))).))) ( -43.40, z-score = -3.20, R) >droEre2.scaffold_4770 10355228 92 + 17746568 -------GCCGCCUCGUCUG----GGCCAUGUUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCGGCAAAAGUUGGCGCUGCCAGC--------GGUCGAGUGUG -------..(((((((.(((----((((((....)))))).((((((.(((((((((((((....))))))))))......))).)))))))--------)).)))).))) ( -43.40, z-score = -2.85, R) >droAna3.scaffold_13340 5899734 100 + 23697760 -------GCCGCCUCGUCUG----GGCCAUGUUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCAGCAAAAGUUUGCGCUGCCAGCUCGCUACCAAUCGACUGCA -------...((.(((..((----((((((....)))))..((((((.(((..((..((((((....))))))....))..))).)))))).......)))..)))..)). ( -36.80, z-score = -1.05, R) >droPer1.super_3 3528070 92 + 7375914 -------GUCGCCUCGUCUA----GGCCAUGUUUGUGGCUUUUGGUAUUGUUUGCCAGCUGACAACAGUCGGCAAAAGUUUAUGGCACCAGC--------GCUUCGGUGCG -------...(((......(----((((((....))))))).(((((.....)))))((((((....))))))..........)))....((--------((....)))). ( -35.30, z-score = -2.49, R) >dp4.chr2 20775831 92 + 30794189 -------GUCGCCUCGUCUA----GGCCAUGUUUGUGGCUUUUGGUAUUGUUUGCCAGCUGACAACAGUCGGCAAAAGUUUAUGGCACCAGC--------GCUUCGGUGCG -------...(((......(----((((((....))))))).(((((.....)))))((((((....))))))..........)))....((--------((....)))). ( -35.30, z-score = -2.49, R) >droWil1.scaffold_181089 8716822 102 + 12369635 ACCGCCUCACUCUUUGUCUGCUAUGCUCAUGUUUGUGGCUUCUGCCAUUGUUUGCCAGCUGACAACAGUCAGCAAAAGUUUAUGCUGCCAGC--------GGUUGGGUUG- (((((........(((((.(((..((........(((((....))))).....)).))).)))))..(.(((((........))))).).))--------))).......- ( -29.52, z-score = -0.20, R) >droVir3.scaffold_13047 217180 80 - 19223366 ---------GCCUUCUCGCC---UGGCCAAGUU-GUGGCUUU-GCCAUUGUUUGCCAGCUGACAACAGUCAGCAAAAGUUUAUGCCGAGAGGCG----------------- ---------((((.((((.(---((((.(((..-(((((...-)))))..)))))))((((((....))))))..........).)))))))).----------------- ( -34.40, z-score = -3.71, R) >droMoj3.scaffold_6540 14686081 86 + 34148556 ---GCCGCACCAUUCUUGUC---UGGCCUUGUU-GUGGCUUU-GCCAUUGUUUGCCAGCUGACAACAGUCAGCAAAAGUUUAUGCCGAGGGGCG----------------- ---(((.(............---((((......-(((((...-))))).....))))((((((....))))))...............).))).----------------- ( -28.00, z-score = -0.47, R) >droGri2.scaffold_14906 2157868 88 - 14172833 GCCGCUUGAGCCUUCUUGUG---UAACCAUGUU-GUGGCUU--GCCAUUGUUUGCCAGCUGACAACAGCCGGCAAAAGUUUAUGCCGAGAGGCG----------------- (((.((((.((.....((.(---(((((((...-)))).))--))))...((((((.((((....)))).)))))).......)))))).))).----------------- ( -31.50, z-score = -1.94, R) >consensus _______GCCGCCUCGUCUG____GGCCAUGUUUGUGGCUUUUGGCAUUGUUUGCCAGCUGACAACAGUCAGCAAAAGUUUACGCUGCCAGC________GGUCGAGUGUG ........................((((((....)))))).........((..((..((((((....))))))....))..))............................ (-15.29 = -14.81 + -0.48)

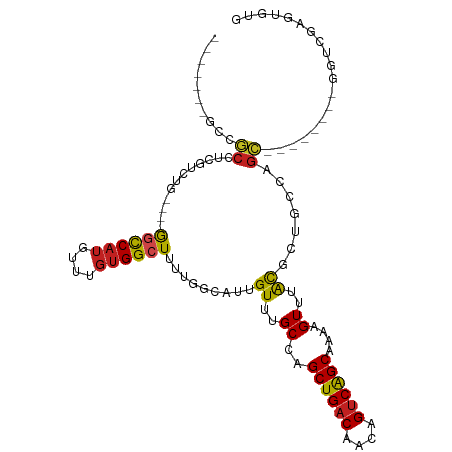
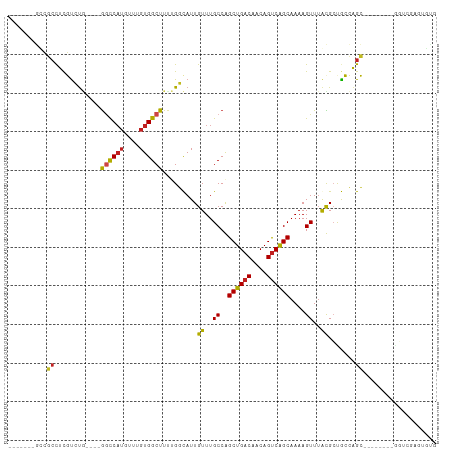
| Location | 10,818,189 – 10,818,281 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.77 |
| Shannon entropy | 0.48397 |
| G+C content | 0.55373 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -16.42 |
| Energy contribution | -15.86 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10818189 92 - 27905053 CACACUCGACC--------GCUGGCAGCGCAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAACAUGGCC----CAGACGAGGCGGC------- ....((((...--------(((((((((((((....)))).....))))))))).((((.....))))....((((......)))).----....)))).....------- ( -30.30, z-score = -1.12, R) >droSim1.chr3R 16878920 92 + 27517382 CACACUCGACC--------GCUGGCAGCGCAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAACAUGGCC----CAGACGAGGCGGC------- ....((((...--------(((((((((((((....)))).....))))))))).((((.....))))....((((......)))).----....)))).....------- ( -30.30, z-score = -1.12, R) >droSec1.super_0 10002033 92 + 21120651 CACACUCGACC--------GCUGGCAGCGCAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAACAUGGCC----CAGACGAGGCGGC------- ....((((...--------(((((((((((((....)))).....))))))))).((((.....))))....((((......)))).----....)))).....------- ( -30.30, z-score = -1.12, R) >droYak2.chr3R 13225515 92 + 28832112 CACACUCGACC--------GCUGGCAGCGCAAACUUUUGCCGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAACAUGGCC----CAGACGAGGCGGC------- ....((((...--------(((((((((((((....)))).....))))))))).((((.....))))....((((......)))).----....)))).....------- ( -30.30, z-score = -1.16, R) >droEre2.scaffold_4770 10355228 92 - 17746568 CACACUCGACC--------GCUGGCAGCGCCAACUUUUGCCGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAACAUGGCC----CAGACGAGGCGGC------- .........((--------(((((((..(......(((((((.(((....))).))))))))..))))....((((......)))).----.......))))).------- ( -29.70, z-score = -0.87, R) >droAna3.scaffold_13340 5899734 100 - 23697760 UGCAGUCGAUUGGUAGCGAGCUGGCAGCGCAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAACAUGGCC----CAGACGAGGCGGC------- .((.(((..((((((....((((((...((((....))))((((....))))))))))......))))))..((((......)))).----..)))...))...------- ( -35.30, z-score = -0.80, R) >droPer1.super_3 3528070 92 - 7375914 CGCACCGAAGC--------GCUGGUGCCAUAAACUUUUGCCGACUGUUGUCAGCUGGCAAACAAUACCAAAAGCCACAAACAUGGCC----UAGACGAGGCGAC------- (((..((....--------.((((.(((((........((.(((....))).))((((..............)))).....))))))----))).))..)))..------- ( -26.64, z-score = -1.20, R) >dp4.chr2 20775831 92 - 30794189 CGCACCGAAGC--------GCUGGUGCCAUAAACUUUUGCCGACUGUUGUCAGCUGGCAAACAAUACCAAAAGCCACAAACAUGGCC----UAGACGAGGCGAC------- (((..((....--------.((((.(((((........((.(((....))).))((((..............)))).....))))))----))).))..)))..------- ( -26.64, z-score = -1.20, R) >droWil1.scaffold_181089 8716822 102 - 12369635 -CAACCCAACC--------GCUGGCAGCAUAAACUUUUGCUGACUGUUGUCAGCUGGCAAACAAUGGCAGAAGCCACAAACAUGAGCAUAGCAGACAAAGAGUGAGGCGGU -.......(((--------(((..(((((........)))))(((.(((((.((((((......((((....))))((....)).)).)))).)))))..)))..)))))) ( -33.60, z-score = -1.79, R) >droVir3.scaffold_13047 217180 80 + 19223366 -----------------CGCCUCUCGGCAUAAACUUUUGCUGACUGUUGUCAGCUGGCAAACAAUGGC-AAAGCCAC-AACUUGGCCA---GGCGAGAAGGC--------- -----------------.((((((((.(..........((((((....))))))((((......((((-...)))).-......))))---).)))).))))--------- ( -33.22, z-score = -3.32, R) >droMoj3.scaffold_6540 14686081 86 - 34148556 -----------------CGCCCCUCGGCAUAAACUUUUGCUGACUGUUGUCAGCUGGCAAACAAUGGC-AAAGCCAC-AACAAGGCCA---GACAAGAAUGGUGCGGC--- -----------------(((.((((((((........)))))).(.(((((...((((......((((-...)))).-......))))---))))).)..)).)))..--- ( -27.22, z-score = -0.72, R) >droGri2.scaffold_14906 2157868 88 + 14172833 -----------------CGCCUCUCGGCAUAAACUUUUGCCGGCUGUUGUCAGCUGGCAAACAAUGGC--AAGCCAC-AACAUGGUUA---CACAAGAAGGCUCAAGCGGC -----------------.((((.(((.(((.....(((((((((((....)))))))))))..))).)--.(((((.-....))))).---.....))))))......... ( -30.40, z-score = -1.97, R) >consensus CACACUCGACC________GCUGGCAGCACAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAACAUGGCC____CAGACGAGGCGGC_______ ...................(((.............(((((((.(((....))).)))))))...........((((......))))............))).......... (-16.42 = -15.86 + -0.56)

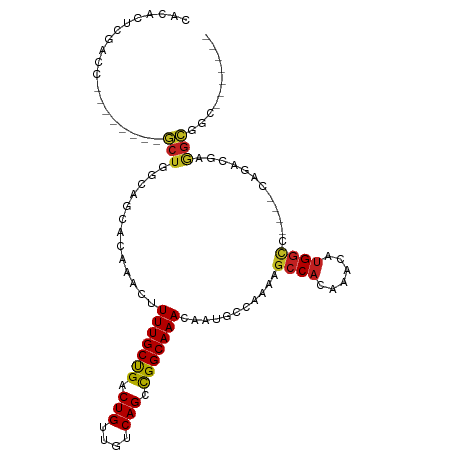
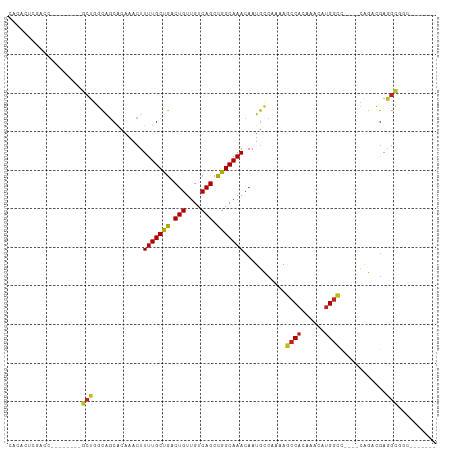
| Location | 10,818,209 – 10,818,304 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.53 |
| Shannon entropy | 0.60339 |
| G+C content | 0.52371 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -14.17 |
| Energy contribution | -13.69 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
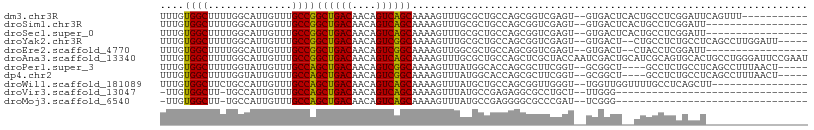
>dm3.chr3R 10818209 95 + 27905053 UUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCAGCAAAAGUUUGCGCUGCCAGCGGUCGAGU--GUGACUCACUGCCUCGGAUUCAGUUU----------- ...(..((((((((((.....)))).((((((....))))))))))).)..)((((((.(((((.(((.--....))))))))...))...))))..----------- ( -33.00, z-score = -0.98, R) >droSim1.chr3R 16878940 89 - 27517382 UUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCAGCAAAAGUUUGCGCUGCCAGCGGUCGAGU--GUGACUCACUGCCUCGGAUU----------------- ((((.(((..((((((.(((..((..((((((....))))))....))..))).)))))).(((.(((.--....))))))))).))))..----------------- ( -32.30, z-score = -1.11, R) >droSec1.super_0 10002053 89 - 21120651 UUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCAGCAAAAGUUUGCGCUGCCAGCGGUCGAGU--GUGACUCACUGCCUCGGAUU----------------- ((((.(((..((((((.(((..((..((((((....))))))....))..))).)))))).(((.(((.--....))))))))).))))..----------------- ( -32.30, z-score = -1.11, R) >droYak2.chr3R 13225535 99 - 28832112 UUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCGGCAAAAGUUUGCGCUGCCAGCGGUCGAGU--GUGACU--CUGCCUCUGCCUCAGCCUUGGAUU----- .(((.(((..((((((.(((((((((((((....))))))))))......))).)))))).(((.(((.--....))--).)))...))).))).........----- ( -37.80, z-score = -1.44, R) >droEre2.scaffold_4770 10355248 87 + 17746568 UUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCGGCAAAAGUUGGCGCUGCCAGCGGUCGAGU--GUGACU--CUACCUCGGAUU----------------- ...((.((.((((((....(((((((((((....))))))))))).((((((...)))))).)))))).--)).)).--............----------------- ( -34.90, z-score = -2.25, R) >droAna3.scaffold_13340 5899754 108 + 23697760 UUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCAGCAAAAGUUUGCGCUGCCAGCUCGCUACCAAUCGACUGCAUCGCAGUGCACUGCCUGGGAUUCCGAAU .....(((...(((((.....)))))((((((....)))))).......(((((((((((.(((........))))))....))))))))..)))(((....)))... ( -36.70, z-score = -0.64, R) >droPer1.super_3 3528090 97 + 7375914 UUUGUGGCUUUUGGUAUUGUUUGCCAGCUGACAACAGUCGGCAAAAGUUUAUGGCACCAGCGCUUCGGU--GCGGCU----GCCUCUGCCUCAGCCUUUAACU----- .....((((...((((...((((((.((((....)))).)))))).......((((((.((((....))--)))).)----)))..))))..)))).......----- ( -39.10, z-score = -2.87, R) >dp4.chr2 20775851 97 + 30794189 UUUGUGGCUUUUGGUAUUGUUUGCCAGCUGACAACAGUCGGCAAAAGUUUAUGGCACCAGCGCUUCGGU--GCGGCU----GCCUCUGCCUCAGCCUUUAACU----- .....((((...((((...((((((.((((....)))).)))))).......((((((.((((....))--)))).)----)))..))))..)))).......----- ( -39.10, z-score = -2.87, R) >droWil1.scaffold_181089 8716853 90 + 12369635 UUUGUGGCUUCUGCCAUUGUUUGCCAGCUGACAACAGUCAGCAAAAGUUUAUGCUGCCAGCGGUUGGGU--UGGUUGGUUUUGCCUCAGCUU---------------- ...(((((....))))).........((((((....))))))..(((((...((.((((((.(......--).))))))...))...)))))---------------- ( -28.70, z-score = -0.14, R) >droVir3.scaffold_13047 217199 72 - 19223366 -UUGUGGCUU-UGCCAUUGUUUGCCAGCUGACAACAGUCAGCAAAAGUUUAUGCCGAGAGGCGCCUGCU--UUGGG-------------------------------- -..(((((..-.)))))......((.((((((....)))))).(((((...((((....))))...)))--)))).-------------------------------- ( -27.10, z-score = -1.85, R) >droMoj3.scaffold_6540 14686106 72 + 34148556 -UUGUGGCUU-UGCCAUUGUUUGCCAGCUGACAACAGUCAGCAAAAGUUUAUGCCGAGGGGCGCCCGAU--UCGGG-------------------------------- -...((((..-.((....))..))))((((((....))))))..........(((....))).((((..--.))))-------------------------------- ( -23.30, z-score = -0.55, R) >consensus UUUGUGGCUUUUGGCAUUGUUUGCCGGCUGACAACAGUCAGCAAAAGUUUGCGCUGCCAGCGGUCGAGU__GUGACU__CUGCCUCGGACU_________________ ....((((..............))))((((((....)))))).................................................................. (-14.17 = -13.69 + -0.48)
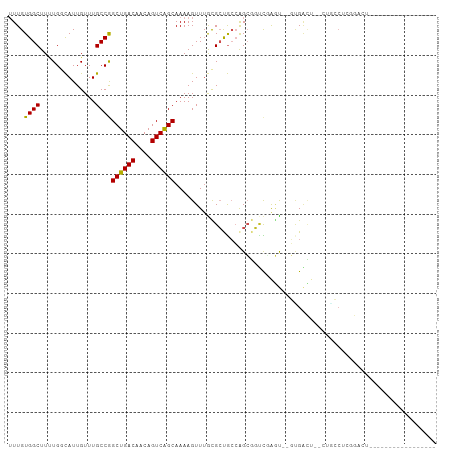
| Location | 10,818,209 – 10,818,304 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.53 |
| Shannon entropy | 0.60339 |
| G+C content | 0.52371 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -13.71 |
| Energy contribution | -12.80 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.710686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
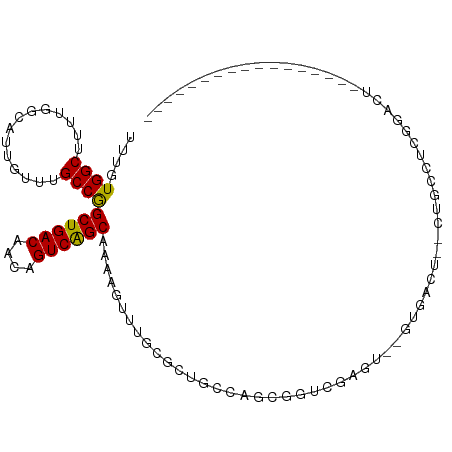
>dm3.chr3R 10818209 95 - 27905053 -----------AAACUGAAUCCGAGGCAGUGAGUCAC--ACUCGACCGCUGGCAGCGCAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAA -----------.............(((((((.(((..--....)))))))(((...((((....))))((((....)))))))((((.....))))....)))..... ( -29.50, z-score = -1.33, R) >droSim1.chr3R 16878940 89 + 27517382 -----------------AAUCCGAGGCAGUGAGUCAC--ACUCGACCGCUGGCAGCGCAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAA -----------------.......(((((((.(((..--....)))))))(((...((((....))))((((....)))))))((((.....))))....)))..... ( -29.50, z-score = -1.67, R) >droSec1.super_0 10002053 89 + 21120651 -----------------AAUCCGAGGCAGUGAGUCAC--ACUCGACCGCUGGCAGCGCAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAA -----------------.......(((((((.(((..--....)))))))(((...((((....))))((((....)))))))((((.....))))....)))..... ( -29.50, z-score = -1.67, R) >droYak2.chr3R 13225535 99 + 28832112 -----AAUCCAAGGCUGAGGCAGAGGCAG--AGUCAC--ACUCGACCGCUGGCAGCGCAAACUUUUGCCGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAA -----.......((((..((((..((..(--((....--.)))..))((((((.(.((((....)))))(((....))).))))))......))))...))))..... ( -38.40, z-score = -2.93, R) >droEre2.scaffold_4770 10355248 87 - 17746568 -----------------AAUCCGAGGUAG--AGUCAC--ACUCGACCGCUGGCAGCGCCAACUUUUGCCGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAA -----------------.......(((.(--((....--.))).)))(((((((..(......(((((((.(((....))).))))))))..))))...)))...... ( -25.50, z-score = -1.18, R) >droAna3.scaffold_13340 5899754 108 - 23697760 AUUCGGAAUCCCAGGCAGUGCACUGCGAUGCAGUCGAUUGGUAGCGAGCUGGCAGCGCAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAA ....((....)).(((..((.(((((...))))))).((((((....((((((...((((....))))((((....))))))))))......))))))..)))..... ( -36.00, z-score = -0.38, R) >droPer1.super_3 3528090 97 - 7375914 -----AGUUAAAGGCUGAGGCAGAGGC----AGCCGC--ACCGAAGCGCUGGUGCCAUAAACUUUUGCCGACUGUUGUCAGCUGGCAAACAAUACCAAAAGCCACAAA -----.......((((..((....(((----(.((((--......)))..).)))).......(((((((.(((....))).))))))).....))...))))..... ( -30.40, z-score = -0.57, R) >dp4.chr2 20775851 97 - 30794189 -----AGUUAAAGGCUGAGGCAGAGGC----AGCCGC--ACCGAAGCGCUGGUGCCAUAAACUUUUGCCGACUGUUGUCAGCUGGCAAACAAUACCAAAAGCCACAAA -----.......((((..((....(((----(.((((--......)))..).)))).......(((((((.(((....))).))))))).....))...))))..... ( -30.40, z-score = -0.57, R) >droWil1.scaffold_181089 8716853 90 - 12369635 ----------------AAGCUGAGGCAAAACCAACCA--ACCCAACCGCUGGCAGCAUAAACUUUUGCUGACUGUUGUCAGCUGGCAAACAAUGGCAGAAGCCACAAA ----------------.((((((((.....))...((--(((((.....)))(((((........)))))...)))))))))).........((((....)))).... ( -24.00, z-score = -0.29, R) >droVir3.scaffold_13047 217199 72 + 19223366 --------------------------------CCCAA--AGCAGGCGCCUCUCGGCAUAAACUUUUGCUGACUGUUGUCAGCUGGCAAACAAUGGCA-AAGCCACAA- --------------------------------.....--....((((((....)))..........((((((....))))))((.((.....)).))-..)))....- ( -21.20, z-score = -0.69, R) >droMoj3.scaffold_6540 14686106 72 - 34148556 --------------------------------CCCGA--AUCGGGCGCCCCUCGGCAUAAACUUUUGCUGACUGUUGUCAGCUGGCAAACAAUGGCA-AAGCCACAA- --------------------------------((((.--..)))).(((....)))..........((((((....))))))((((...........-..))))...- ( -21.12, z-score = -0.64, R) >consensus _________________AAGCAGAGGCAG__AGUCAC__ACUCGACCGCUGGCAGCGCAAACUUUUGCUGACUGUUGUCAGCCGGCAAACAAUGCCAAAAGCCACAAA .................................................(((((.........(((((((.(((....))).)))))))...)))))........... (-13.71 = -12.80 + -0.91)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:20 2011