| Sequence ID | dm3.chr3R |
|---|---|
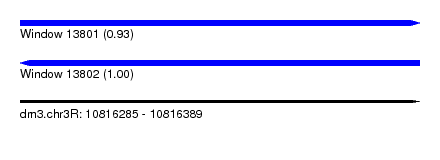
| Location | 10,816,285 – 10,816,389 |
| Length | 104 |
| Max. P | 0.999859 |

| Location | 10,816,285 – 10,816,389 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.52998 |
| G+C content | 0.61304 |
| Mean single sequence MFE | -41.69 |
| Consensus MFE | -16.97 |
| Energy contribution | -18.33 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
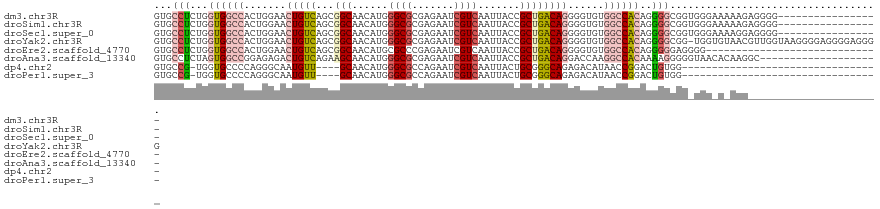
>dm3.chr3R 10816285 104 + 27905053 GUGCCUCUGGUGGCCACUGGAACUGUCAGCGGCAACAUGGGCGCGAGAAUCGUCAAUUACCGCUGACAGGGGUGUGGCCACAGGGGCGGUGGGAAAAAGAGGGG----------------- .((((((((.(((((((.....((((((((((.......((((.......)))).....))))))))))....)))))))))))))))................----------------- ( -50.60, z-score = -4.22, R) >droSim1.chr3R 16875305 104 - 27517382 GUGCCUCUGGUGGCCACUGGAACUGUCAGCGGCAACAUGGGCGCGAGAAUCGUCAAUUACCGCUGACAGGGGUGUGGCCACAGGGGCGGUGGGAAAAAGAGGGG----------------- .((((((((.(((((((.....((((((((((.......((((.......)))).....))))))))))....)))))))))))))))................----------------- ( -50.60, z-score = -4.22, R) >droSec1.super_0 10000152 104 - 21120651 GUGCCUCUGGUGGCCACUGGAACUGUCAGCGGCAACAUGGGCGCGAGAAUCGUCAAUUACCGCUGACAGGGGUGUGGCCACAGGGGCGGUGGGAAAAGGAGGGG----------------- .((((((((.(((((((.....((((((((((.......((((.......)))).....))))))))))....)))))))))))))))................----------------- ( -50.60, z-score = -4.02, R) >droYak2.chr3R 13223557 120 - 28832112 GUGCCUCUGGUGGCCACUGGAACUGUCAGCGGCAACAUGGGCGCGAGAAUCGUCAAUUACCGCUGACAGGGGUGUGGCCACAGGGGCGG-UGGUGUAACGUUGGUAAGGGGAGGGGAGGGG .((((((((.(((((((.....((((((((((.......((((.......)))).....))))))))))....))))))))))))))).-............................... ( -50.60, z-score = -2.32, R) >droEre2.scaffold_4770 10353347 92 + 17746568 GUGCCUCUGGUGGCCACUGGAACUGUCAGCGGCAACAUGCGCCCGAGAAUCGUCAAUUACCGCUGACAGGGGUGUGGCCACAGGGGGAGGGG----------------------------- ...(((((.((((((((.....((((((((((......(((.........)))......))))))))))....)))))))).))))).....----------------------------- ( -44.40, z-score = -2.53, R) >droAna3.scaffold_13340 5896040 101 + 23697760 GUGCCUCUAGUGGCCGGAGAGACUGUCAGAAGCAACAUGGGCGCGAGAAUCGUCAAUUACCGCUGACAGGACCAAGGCCACAAAAGGGGGUAACACAAGGC-------------------- .(((((((.((((((.......(((((((..........((((.......))))........)))))))......))))))....))))))).........-------------------- ( -34.29, z-score = -1.88, R) >dp4.chr2 20773710 83 + 30794189 GUGCCG-UGGUGCCCCAGGGCAAUGUU----GCAACAUGGGCGCCAGAAUCGUCAAUUACUGCGGGCAGAGACAUAACCGGACUGUGG--------------------------------- .((((.-((((((((....(((....)----)).....))))))))....(((........)))))))....((((.......)))).--------------------------------- ( -26.20, z-score = 0.67, R) >droPer1.super_3 3526186 83 + 7375914 GUGCCG-UGGUGCCCCAGGGCAAUGUU----GCAACAUGGGCGCCAGAAUCGUCAAUUACUGCGGGCAGAGACAUAACCGGACUGUGG--------------------------------- .((((.-((((((((....(((....)----)).....))))))))....(((........)))))))....((((.......)))).--------------------------------- ( -26.20, z-score = 0.67, R) >consensus GUGCCUCUGGUGGCCACUGGAACUGUCAGCGGCAACAUGGGCGCGAGAAUCGUCAAUUACCGCUGACAGGGGUGUGGCCACAGGGGCGG_GGG_A_AAG______________________ ...(((...((((((.......(((((((..........((((.......))))........)))))))......))))))..)))................................... (-16.97 = -18.33 + 1.36)
| Location | 10,816,285 – 10,816,389 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 70.52 |
| Shannon entropy | 0.52998 |
| G+C content | 0.61304 |
| Mean single sequence MFE | -39.67 |
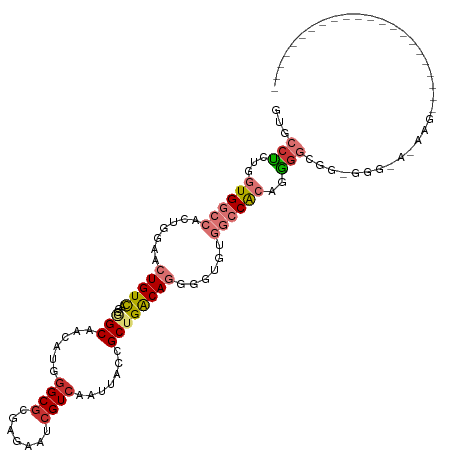
| Consensus MFE | -18.68 |
| Energy contribution | -20.92 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.24 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.61 |
| SVM RNA-class probability | 0.999859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10816285 104 - 27905053 -----------------CCCCUCUUUUUCCCACCGCCCCUGUGGCCACACCCCUGUCAGCGGUAAUUGACGAUUCUCGCGCCCAUGUUGCCGCUGACAGUUCCAGUGGCCACCAGAGGCAC -----------------.................(((.((((((((((....((((((((((((((((.((.......))..)).)))))))))))))).....))))))).))).))).. ( -47.40, z-score = -7.01, R) >droSim1.chr3R 16875305 104 + 27517382 -----------------CCCCUCUUUUUCCCACCGCCCCUGUGGCCACACCCCUGUCAGCGGUAAUUGACGAUUCUCGCGCCCAUGUUGCCGCUGACAGUUCCAGUGGCCACCAGAGGCAC -----------------.................(((.((((((((((....((((((((((((((((.((.......))..)).)))))))))))))).....))))))).))).))).. ( -47.40, z-score = -7.01, R) >droSec1.super_0 10000152 104 + 21120651 -----------------CCCCUCCUUUUCCCACCGCCCCUGUGGCCACACCCCUGUCAGCGGUAAUUGACGAUUCUCGCGCCCAUGUUGCCGCUGACAGUUCCAGUGGCCACCAGAGGCAC -----------------.................(((.((((((((((....((((((((((((((((.((.......))..)).)))))))))))))).....))))))).))).))).. ( -47.40, z-score = -7.00, R) >droYak2.chr3R 13223557 120 + 28832112 CCCCUCCCCUCCCCUUACCAACGUUACACCA-CCGCCCCUGUGGCCACACCCCUGUCAGCGGUAAUUGACGAUUCUCGCGCCCAUGUUGCCGCUGACAGUUCCAGUGGCCACCAGAGGCAC ...............................-..(((.((((((((((....((((((((((((((((.((.......))..)).)))))))))))))).....))))))).))).))).. ( -47.40, z-score = -6.43, R) >droEre2.scaffold_4770 10353347 92 - 17746568 -----------------------------CCCCUCCCCCUGUGGCCACACCCCUGUCAGCGGUAAUUGACGAUUCUCGGGCGCAUGUUGCCGCUGACAGUUCCAGUGGCCACCAGAGGCAC -----------------------------..((((.....((((((((....((((((((((((((((.((.....))....)).)))))))))))))).....))))))))..))))... ( -45.10, z-score = -4.88, R) >droAna3.scaffold_13340 5896040 101 - 23697760 --------------------GCCUUGUGUUACCCCCUUUUGUGGCCUUGGUCCUGUCAGCGGUAAUUGACGAUUCUCGCGCCCAUGUUGCUUCUGACAGUCUCUCCGGCCACUAGAGGCAC --------------------..............(((((.((((((..((..(((((((.((((((((.((.......))..)).)))))).)))))))..))...)))))).)))))... ( -35.30, z-score = -2.77, R) >dp4.chr2 20773710 83 - 30794189 ---------------------------------CCACAGUCCGGUUAUGUCUCUGCCCGCAGUAAUUGACGAUUCUGGCGCCCAUGUUGC----AACAUUGCCCUGGGGCACCA-CGGCAC ---------------------------------.....(.(((((((.....(((....)))....)))).....(((.((((.....((----(....)))....)))).)))-)))).. ( -23.70, z-score = 0.58, R) >droPer1.super_3 3526186 83 - 7375914 ---------------------------------CCACAGUCCGGUUAUGUCUCUGCCCGCAGUAAUUGACGAUUCUGGCGCCCAUGUUGC----AACAUUGCCCUGGGGCACCA-CGGCAC ---------------------------------.....(.(((((((.....(((....)))....)))).....(((.((((.....((----(....)))....)))).)))-)))).. ( -23.70, z-score = 0.58, R) >consensus ______________________CUU_U_CCC_CCCCCCCUGUGGCCACACCCCUGUCAGCGGUAAUUGACGAUUCUCGCGCCCAUGUUGCCGCUGACAGUUCCAGUGGCCACCAGAGGCAC ...................................((.((((((((......(((((((.((((((((.((.......))..)).)))))).))))))).......)))))).)).))... (-18.68 = -20.92 + 2.24)
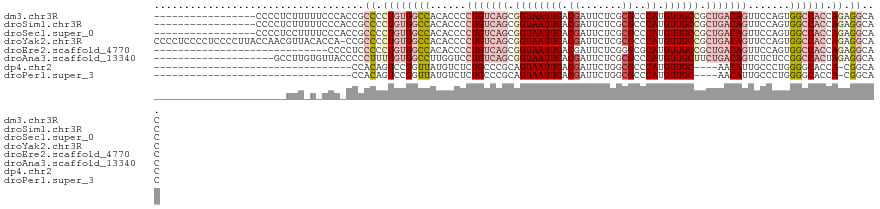

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:17 2011