
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,813,775 – 10,813,870 |
| Length | 95 |
| Max. P | 0.988364 |

| Location | 10,813,775 – 10,813,868 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 59.74 |
| Shannon entropy | 0.83856 |
| G+C content | 0.53049 |
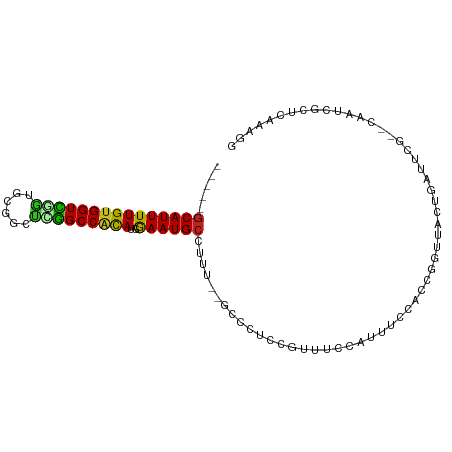
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -16.65 |
| Energy contribution | -15.66 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.34 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
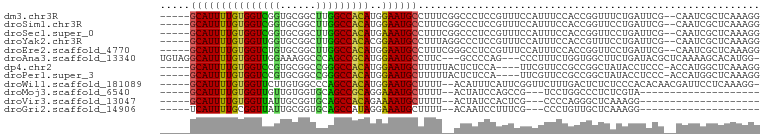
>dm3.chr3R 10813775 93 - 27905053 -----GCAUUUUGUGGUCGGUGCGGCUUGGCCACAUGGAAUGCCUUUCGGCCCUCCGUUUCCAUUUCCACCGGUUUCUGAUUCG--CAAUCGCUCAAAGG -----((...((((((((((((.((..(((....(((((..(((....)))..)))))..)))..))))))))........)))--)))..))....... ( -26.90, z-score = -0.31, R) >droSim1.chr3R 16872787 93 + 27517382 -----GCAUUUUGUGGUCGGUGCGGCUUGGCCACAUGGAAUGCCUUUCGGCCCUCCGUUUCCAUUUCCACCGGUUCCUGAUUCG--CAAUCGCUCAAAGG -----((...((((((((((((.((..(((....(((((..(((....)))..)))))..)))..))))))))........)))--)))..))....... ( -26.90, z-score = -0.16, R) >droSec1.super_0 9997622 93 + 21120651 -----GCAUUUUGUGGUCGGUGCGGCUUGGCCACAUGAAAUGCCUUUCGGCCCUCCGUUUCCAUUUCCACCGGUUCCUGAUUCG--CAAUCGCUCAAAGG -----(((((((((((((((......)))))))))..))))))((((.(((....((..((.....((...)).....))..))--.....))).)))). ( -22.90, z-score = 0.69, R) >droYak2.chr3R 13220960 93 + 28832112 -----GCAUUUUGUGGUUGGUGCGGCUUGGCCACACGGAAUGCCUUUAGGCCCUCCGUUUCCAUUUCCACCGUUUCCUGAUUCG--CAAUCGCUCAAAGG -----((...((((((.(((((.((..(((....(((((..(((....)))..)))))..)))..))))))).........)))--)))..))....... ( -26.70, z-score = -0.64, R) >droEre2.scaffold_4770 10350797 93 - 17746568 -----GCAUUUUGUGGUCUGUGCGGCUUGGCCACAUGGAAUGCCUUUCGGGCCUCCGUUUCCAUUUCCACCGGUUCCUGAUUCG--CAAUCGCUCAAAGG -----((....(((((((..........)))))))..((.(((...(((((...(((.............)))..)))))...)--)).))))....... ( -24.92, z-score = 0.34, R) >droAna3.scaffold_13340 5893575 93 - 23697760 UGUAGGCAUUUUGUGGUUGGAAAGGCCCAGCCGCAUGGAAUGCCUUC---GCCCCAG---CCCUUUCUGGUGGCUUCUGAUACGCUCAAAAGCACAUGG- .(.(((((((((((((((((......)))))))))..)))))))).)---(((((((---......)))).))).........(((....)))......- ( -37.30, z-score = -2.18, R) >dp4.chr2 20771084 90 - 30794189 -----GCAUUUUGUGGUCCGUGCGGCCGGGCCACAUGGAAUGCUUUUUACUCUCCA----UUCGUUCCGCCGGCUAUACCUCCC-ACCAUGGCUCAAAGG -----(((((((((((((((......)))))))))..)))))).............----......((...((((((.......-...))))))....)) ( -25.80, z-score = -0.37, R) >droPer1.super_3 3523621 90 - 7375914 -----GCAUUUUGUGGUCCGUGCGGCCGGGCCACAUGGAAUGCUUUUUACUCUCCA----UUCGUUCCGCCGGCUAUACCUCCC-ACCAUGGCUCAAAGG -----(((((((((((((((......)))))))))..)))))).............----......((...((((((.......-...))))))....)) ( -25.80, z-score = -0.37, R) >droWil1.scaffold_181089 8711503 92 - 12369635 -----GCAUUUUGUGGUUCUUGUGGCCCAGCCACAUGGAAUGCUUUU--ACAUUUCAUUCGGUUCUUUGACUCUCUCCCACACAACGAUUCCUCAAAGG- -----...(((((.((.(((((((....((((..((((((((.....--.))))))))..))))....((.....))...))))).))..)).))))).- ( -19.40, z-score = -0.86, R) >droMoj3.scaffold_6540 14680333 70 - 34148556 -----GCAUUUUGUGGUUGUUGUGGUGCAGCCGCAGGAAAUGCUUUU--ACUAUCCAGCCG---UCCUGGCCCUCUCGUA-------------------- -----(((((((((((((((......)))))))))..))))))....--........(((.---....))).........-------------------- ( -21.40, z-score = -1.19, R) >droVir3.scaffold_13047 210394 70 + 19223366 -----GCAUUUUGUGGUUAUUGCGGUGCAGCCACAGAAAAUGCUUUU--ACUAUCCACUCG---CCCCAGGGCUCAAAGG-------------------- -----((((((((((((...(((...)))))))))))..))))....--...........(---((....))).......-------------------- ( -19.20, z-score = -0.75, R) >droGri2.scaffold_14906 2151918 70 + 14172833 -----UCAUUUUGCGGUUAUUGCGGUGCAGCCAUAGGAAAUGCUUUU--ACAAUCCUUUCG---CCCUGUUGCUCAAAGG-------------------- -----...(((((.(((....((((.((......((((..((.....--.)).))))...)---).)))).)))))))).-------------------- ( -10.80, z-score = 1.77, R) >consensus _____GCAUUUUGUGGUCGGUGCGGCUCGGCCACAUGGAAUGCCUUU__GCCCUCCGUUUCCAUUUCCACCGGUUACUGAUUCG__CAAUCGCUCAAAGG .....(((((((((((((((......)))))))))..))))))......................................................... (-16.65 = -15.66 + -0.99)
| Location | 10,813,777 – 10,813,870 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 59.54 |
| Shannon entropy | 0.80429 |
| G+C content | 0.52206 |
| Mean single sequence MFE | -25.51 |
| Consensus MFE | -16.35 |
| Energy contribution | -15.36 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.988364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
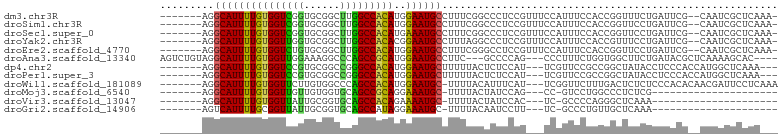
>dm3.chr3R 10813777 93 - 27905053 -------AGGCAUUUUGUGGUCGGUGCGGCUUGGCCACAUGGAAUGCCUUUCGGCCCUCCGUUUCCAUUUCCACCGGUUUCUGAUUCG--CAAUCGCUCAAA- -------.(((...((((((((((((.((..(((....(((((..(((....)))..)))))..)))..))))))))........)))--)))..)))....- ( -27.70, z-score = -0.38, R) >droSim1.chr3R 16872789 93 + 27517382 -------AGGCAUUUUGUGGUCGGUGCGGCUUGGCCACAUGGAAUGCCUUUCGGCCCUCCGUUUCCAUUUCCACCGGUUCCUGAUUCG--CAAUCGCUCAAA- -------.(((...((((((((((((.((..(((....(((((..(((....)))..)))))..)))..))))))))........)))--)))..)))....- ( -27.70, z-score = -0.22, R) >droSec1.super_0 9997624 93 + 21120651 -------AGGCAUUUUGUGGUCGGUGCGGCUUGGCCACAUGAAAUGCCUUUCGGCCCUCCGUUUCCAUUUCCACCGGUUCCUGAUUCG--CAAUCGCUCAAA- -------(((((((((((((((((......)))))))))..)))))))).((((....(((.............)))...))))...(--(....)).....- ( -25.02, z-score = 0.17, R) >droYak2.chr3R 13220962 93 + 28832112 -------AGGCAUUUUGUGGUUGGUGCGGCUUGGCCACACGGAAUGCCUUUAGGCCCUCCGUUUCCAUUUCCACCGUUUCCUGAUUCG--CAAUCGCUCAAA- -------.(((...((((((.(((((.((..(((....(((((..(((....)))..)))))..)))..))))))).........)))--)))..)))....- ( -27.50, z-score = -0.73, R) >droEre2.scaffold_4770 10350799 93 - 17746568 -------AGGCAUUUUGUGGUCUGUGCGGCUUGGCCACAUGGAAUGCCUUUCGGGCCUCCGUUUCCAUUUCCACCGGUUCCUGAUUCG--CAAUCGCUCAAA- -------(((((((((((((((..........)))))))..)))))))).(((((...(((.............)))..)))))...(--(....)).....- ( -27.82, z-score = -0.42, R) >droAna3.scaffold_13340 5893579 93 - 23697760 AGUCUGUAGGCAUUUUGUGGUUGGAAAGGCCCAGCCGCAUGGAAUGCCUUC---GCCCCAG---CCCUUUCUGGUGGCUUCUGAUACGCUCAAAAGCAC---- .(((.(.(((((((((((((((((......)))))))))..)))))))).)---(((((((---......)))).)))....)))..(((....)))..---- ( -38.20, z-score = -2.79, R) >dp4.chr2 20771086 90 - 30794189 -------AGGCAUUUUGUGGUCCGUGCGGCCGGGCCACAUGGAAUGCUUUUUACUCUCCAU---UCGUUCCGCCGGCUAUACCUCCCACCAUGGCUCAAA--- -------.(((....(((((((((......))))))))).((((((...............---.)))))))))((((((..........))))))....--- ( -29.29, z-score = -1.29, R) >droPer1.super_3 3523623 90 - 7375914 -------AGGCAUUUUGUGGUCCGUGCGGCCGGGCCACAUGGAAUGCUUUUUACUCUCCAU---UCGUUCCGCCGGCUAUACCUCCCACCAUGGCUCAAA--- -------.(((....(((((((((......))))))))).((((((...............---.)))))))))((((((..........))))))....--- ( -29.29, z-score = -1.29, R) >droWil1.scaffold_181089 8711505 92 - 12369635 -------AGGCAUUUUGUGGUUCUUGUGGCCCAGCCACAUGGAAUGC-UUUUACAUUUCAU---UCGGUUCUUUGACUCUCUCCCACACAACGAUUCCUCAAA -------(((.((((((((.....((((((...))))))(((((((.-.....))))))).---......................))))).))).))).... ( -20.40, z-score = -1.01, R) >droMoj3.scaffold_6540 14680335 70 - 34148556 -------AGGCAUUUUGUGGUUGUUGUGGUGCAGCCGCAGGAAAUGC-UUUUACUAUCCAG---CC-GUCCUGGCCCUCUCG--------------------- -------(((((((((((((((((......)))))))))..))))))-))..........(---((-.....))).......--------------------- ( -22.50, z-score = -1.16, R) >droVir3.scaffold_13047 210396 70 + 19223366 -------AGGCAUUUUGUGGUUAUUGCGGUGCAGCCACAGAAAAUGC-UUUUACUAUCCAC---UC-GCCCCAGGGCUCAAA--------------------- -------((((((((((((((...(((...)))))))))))..))))-))...........---..-(((....))).....--------------------- ( -20.30, z-score = -0.93, R) >droGri2.scaffold_14906 2151920 70 + 14172833 -------AGUCAUUUUGCGGUUAUUGCGGUGCAGCCAUAGGAAAUGC-UUUUACAAUCCUU---UC-GCCCUGUUGCUCAAA--------------------- -------(((.((...((((..((((..(.(((.((...))...)))-..)..))))....---))-))...)).)))....--------------------- ( -10.40, z-score = 1.77, R) >consensus _______AGGCAUUUUGUGGUCGGUGCGGCUCGGCCACAUGGAAUGCCUUUUACCCCCCAG___CCAUUUCCACCGCUUUCUGAUUCG__CAAUCGCUCA___ .........(((((((((((((((......)))))))))..))))))........................................................ (-16.35 = -15.36 + -0.99)
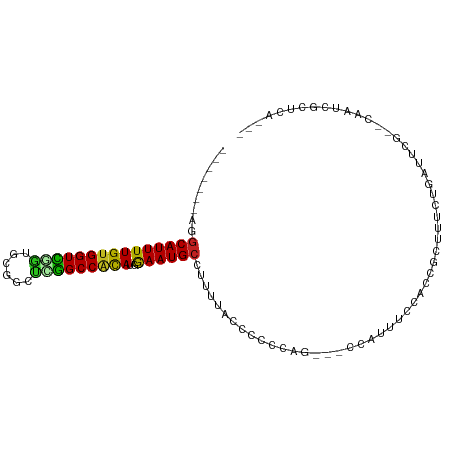
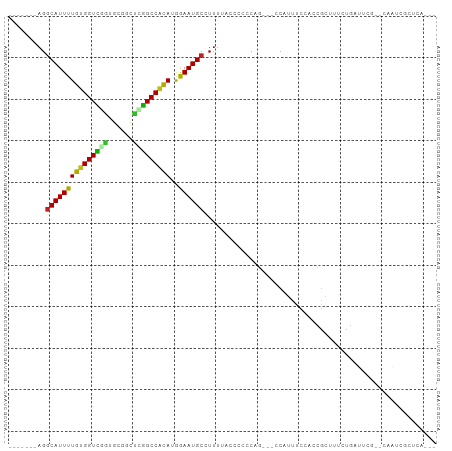
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:15 2011