
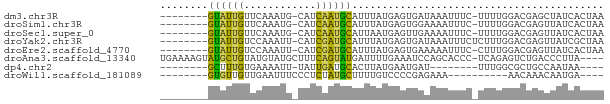
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,793,011 – 10,793,075 |
| Length | 64 |
| Max. P | 0.720014 |

| Location | 10,793,011 – 10,793,075 |
|---|---|
| Length | 64 |
| Sequences | 8 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 58.53 |
| Shannon entropy | 0.79121 |
| G+C content | 0.33800 |
| Mean single sequence MFE | -11.05 |
| Consensus MFE | -3.86 |
| Energy contribution | -3.00 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 2.17 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720014 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
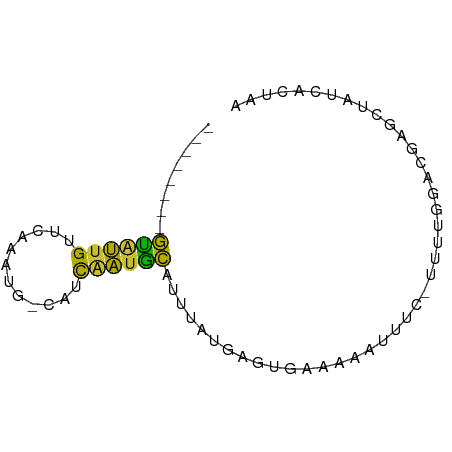
>dm3.chr3R 10793011 64 - 27905053 --------GUAUUGUUCAAAUG-CAUCAAUGCAUUUAUGAGUGAUAAAUUUC-UUUUGGACGAGCUAUCACUAA --------.........(((((-((....)))))))...(((((((..((((-.....)).))..))))))).. ( -12.80, z-score = -1.37, R) >droSim1.chr3R 16852050 64 + 27517382 --------GUAUUGUUCAAAUG-CAUCAAUGCAUUUAUGAGUGGAAAAUUUC-UUUUGGACGAGUUAUCACUAA --------...(((((((((((-((....)))))))..(((........)))-....))))))........... ( -10.80, z-score = -0.52, R) >droSec1.super_0 9977315 64 + 21120651 --------GUAUUGUUCAAAUG-CAUCAAUGCAUUAAUGAGUUGAAAAUUUC-UUUUGGACGAGUUAUCACUAA --------...(((((((((((-((....))))))...(((.(....).)))-...)))))))........... ( -10.90, z-score = -0.71, R) >droYak2.chr3R 13199718 65 + 28832112 --------GUAUUGUCCAAAUU-CAUCGAUGCAUUUAUGAGUGAUAAAUUUCUCUUUGGACGAGUUAUCGCUAA --------((((((........-...)))))).......((((((((.(((.((....)).))))))))))).. ( -12.60, z-score = -1.20, R) >droEre2.scaffold_4770 10329865 64 - 17746568 --------GUAUUGUCCAAAUU-CAUCGAUGCAUUUAUGAGUGAAAAAUUUC-CUUUGGACGAGUUAUCACUAA --------...(((((((((((-(((.((....)).))))))(((....)))-..))))))))........... ( -11.30, z-score = -1.03, R) >droAna3.scaffold_13340 5874024 69 - 23697760 UGAAAAGUAUGCUGUAUGUAUGCUUUCAGUAUGAUUUUGAAAUCCAGCACCC-UCAGAGUCUGACCCUUA---- (((((.((((((.....)))))))))))((..((((((((............-))))))))..)).....---- ( -15.30, z-score = -2.38, R) >dp4.chr2 20746278 53 - 30794189 --------GCUUUGUGAAAAUU-UAUUGAUGCACUUAUGAAUGAU--------UUUGGCGCUGCCAAUAA---- --------((..(((.((((((-.(((.(((....))).))))))--------))).)))..))......---- ( -9.00, z-score = -0.72, R) >droWil1.scaffold_181089 8688663 52 - 12369635 --------GUGUUGUUGAAUUUCCCUCUAUGCUUUUGUCCCCGAGAAA----------AACAAACAAUGA---- --------..((((((...(((..(((...((....))....)))..)----------))..))))))..---- ( -5.70, z-score = -0.88, R) >consensus ________GUAUUGUUCAAAUG_CAUCAAUGCAUUUAUGAGUGAAAAAUUUC_UUUUGGACGAGCUAUCACUAA ........((((((............)))))).......................................... ( -3.86 = -3.00 + -0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:14 2011