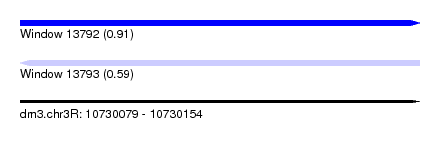
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,730,079 – 10,730,154 |
| Length | 75 |
| Max. P | 0.909307 |

| Location | 10,730,079 – 10,730,154 |
|---|---|
| Length | 75 |
| Sequences | 5 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 73.58 |
| Shannon entropy | 0.47721 |
| G+C content | 0.40172 |
| Mean single sequence MFE | -14.60 |
| Consensus MFE | -8.22 |
| Energy contribution | -9.42 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.909307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
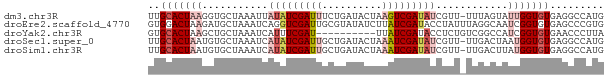
>dm3.chr3R 10730079 75 + 27905053 CAUGGCCUCACACCAAUACUAAA-AACGAUAUCGACUUAGUAUCAGAAAUCGAUAUAAUUUAGCACCUUAGUGCAA ..(((.......))).......(-((..(((((((.((........)).)))))))..))).((((....)))).. ( -11.90, z-score = -1.87, R) >droEre2.scaffold_4770 10266189 76 + 17746568 CACGGGCUCACACCGAUUGCCUAAAUAGGUAUCGAUAAGAUAUACGCAAUCGACCUGAUUUAGCAUCUUAGUCCAC ...(((((.....(((((((((....))(((((.....)))))..)))))))....(((.....)))..))))).. ( -18.00, z-score = -2.09, R) >droYak2.chr3R 13134335 66 - 28832112 UAAGGGUUCACACCGAUGGCCGACAGAGGUAUCGAUAA----------AUCGAAAUGAUUUAGCAGCUUAGUGCAC .....((.(((..(((((.((......))))))).(((----------(((.....))))))........))).)) ( -12.50, z-score = -0.37, R) >droSec1.super_0 9915386 75 - 21120651 CAUGGCCUCACACCAUUAGUCAA-AACGAUAUCGAUUUAGUAUCAGCAAUCGAUAUGAUUUAGCACAUUAGUGCAA .((((.......)))).......-....(((((((((..((....)))))))))))......((((....)))).. ( -14.50, z-score = -1.81, R) >droSim1.chr3R 16786225 75 - 27517382 CAUGGCCUCACACCAUAAGUCAA-AACGAUAUCGAUUUAGUAUCAGCAAUCGAUAUGAUUUAGCACAUUAGUGCAA ..(((.......)))(((((((.-.....((((((((..((....)))))))))))))))))((((....)))).. ( -16.10, z-score = -2.50, R) >consensus CAUGGCCUCACACCAAUAGUCAA_AACGAUAUCGAUUUAGUAUCAGCAAUCGAUAUGAUUUAGCACCUUAGUGCAA ..(((.......))).............(((((((((..........)))))))))......((((....)))).. ( -8.22 = -9.42 + 1.20)

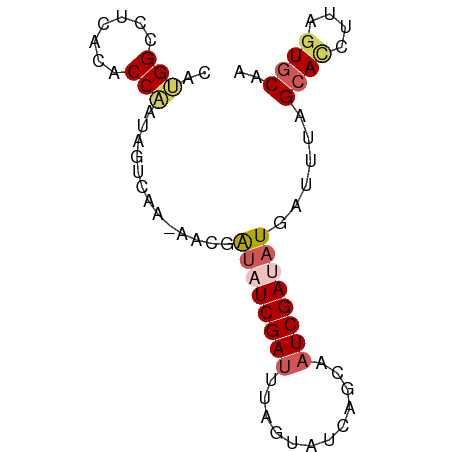
| Location | 10,730,079 – 10,730,154 |
|---|---|
| Length | 75 |
| Sequences | 5 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 73.58 |
| Shannon entropy | 0.47721 |
| G+C content | 0.40172 |
| Mean single sequence MFE | -17.76 |
| Consensus MFE | -7.98 |
| Energy contribution | -10.70 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.591196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10730079 75 - 27905053 UUGCACUAAGGUGCUAAAUUAUAUCGAUUUCUGAUACUAAGUCGAUAUCGUU-UUUAGUAUUGGUGUGAGGCCAUG (..(((((..((((((((..((((((((((........))))))))))....-)))))))))))))..)....... ( -22.20, z-score = -3.35, R) >droEre2.scaffold_4770 10266189 76 - 17746568 GUGGACUAAGAUGCUAAAUCAGGUCGAUUGCGUAUAUCUUAUCGAUACCUAUUUAGGCAAUCGGUGUGAGCCCGUG ((.((((..(((.....))).)))).)).(((....((.(((((((.(((....)))..))))))).))...))). ( -16.30, z-score = 0.19, R) >droYak2.chr3R 13134335 66 + 28832112 GUGCACUAAGCUGCUAAAUCAUUUCGAU----------UUAUCGAUACCUCUGUCGGCCAUCGGUGUGAACCCUUA ..(((......)))....((((.(((((----------...((((((....))))))..))))).))))....... ( -13.30, z-score = -1.04, R) >droSec1.super_0 9915386 75 + 21120651 UUGCACUAAUGUGCUAAAUCAUAUCGAUUGCUGAUACUAAAUCGAUAUCGUU-UUGACUAAUGGUGUGAGGCCAUG ..((((....)))).((((.(((((((((..........))))))))).)))-)......(((((.....))))). ( -18.40, z-score = -2.14, R) >droSim1.chr3R 16786225 75 + 27517382 UUGCACUAAUGUGCUAAAUCAUAUCGAUUGCUGAUACUAAAUCGAUAUCGUU-UUGACUUAUGGUGUGAGGCCAUG ..((((....)))).((((.(((((((((..........))))))))).)))-).....((((((.....)))))) ( -18.60, z-score = -2.13, R) >consensus UUGCACUAAGGUGCUAAAUCAUAUCGAUUGCUGAUACUAAAUCGAUAUCGUU_UUGACUAUUGGUGUGAGGCCAUG ..(((((((.......(((.(((((((((..........))))))))).)))........)))))))......... ( -7.98 = -10.70 + 2.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:09 2011