
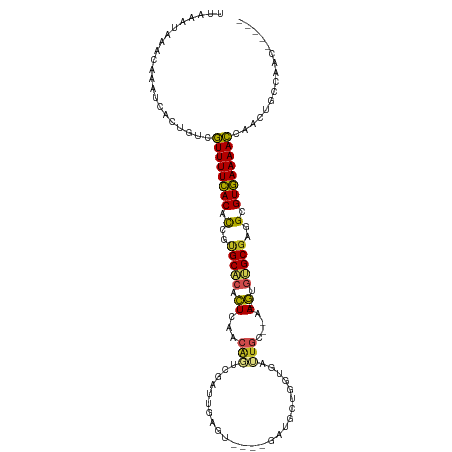
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,728,288 – 10,728,392 |
| Length | 104 |
| Max. P | 0.999943 |

| Location | 10,728,288 – 10,728,392 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 66.72 |
| Shannon entropy | 0.67681 |
| G+C content | 0.42276 |
| Mean single sequence MFE | -32.56 |
| Consensus MFE | -11.60 |
| Energy contribution | -10.68 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.998734 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 10728288 104 + 27905053 UGAAAUAAACAAAUCACGGUCGUUUUCACACCGUGCACACUCCACAGUCGAUUGAGU----GAUGCUGGUGACUGC--GA--GCGUGCGAGGCGUGAAAACCAACUGCCAAC----- ................((((.((((((((.((.(((((.(((..((((((....((.----....))..)))))).--))--).))))).)).))))))))..)))).....----- ( -35.20, z-score = -2.11, R) >droSim1.chr3R 16784464 104 - 27517382 UUAAAUAAACAAAUCACGGUCGUUUUCACACCGUGCACACUCCACAGUCGAUUGAGU----GAUGCUGGUGACUGC--GA--GCGUGCGAGGCGUGAAAACCAACUGCCAAC----- ................((((.((((((((.((.(((((.(((..((((((....((.----....))..)))))).--))--).))))).)).))))))))..)))).....----- ( -35.20, z-score = -2.40, R) >droSec1.super_0 9913591 104 - 21120651 UUAAAUAAACAAAUCACGGUCGUUUUCACACCGUGCACACUCCACAGUCGAUUGAGU----GAUGCUGGUGACUGC--GA--GCGUGCGAGGCGUGAAAACCAACUGCCAAC----- ................((((.((((((((.((.(((((.(((..((((((....((.----....))..)))))).--))--).))))).)).))))))))..)))).....----- ( -35.20, z-score = -2.40, R) >droYak2.chr3R 13132536 104 - 28832112 UUAAAUAAACAAAUCGCCGUCGUUUUCACACCGUGCACACUCCACAGUCGAUUGAGU----GAUGCUGGUGACUGC--AA--GUGUGCGAGGCGUGAAAACCAACUGCCAAC----- ...............((.((.((((((((.((.((((((((...((.((....)).)----).(((.(....).))--))--))))))).)).))))))))..)).))....----- ( -35.80, z-score = -3.02, R) >droEre2.scaffold_4770 10264382 104 + 17746568 UGAAAUAAACAAAUCGCCGCCGUUUUCACACCAUGCACACUCCACAGUCGAUUGAGU----GAUGCUGGUGACUGC--AA--GUGUGCGAGGCGUGAAAACCAACUGCCAAC----- ...............((.(..((((((((.((.((((((((...((.((....)).)----).(((.(....).))--))--))))))).)).))))))))...).))....----- ( -34.30, z-score = -2.71, R) >droAna3.scaffold_13340 5807786 106 + 23697760 UAUAAAAAACAAAACUCAGUCGUUUUCACAUCGGGCACACUCUGCAAUCGAUUGAGU----GAUGCUGGUGAUUGCUAAA--GUGUGCGAGGCGUGAAAACUAACAGCUUAA----- .....................((((((((..(..(((((((..(((((((....((.----....))..)))))))...)--))))))..)..))))))))...........----- ( -32.20, z-score = -2.79, R) >dp4.chr2 20663651 109 + 30794189 UUACAUAAACAAAUCUUUGUCAUUUUUACACCAUGCACACUCAGCAAUCAAUUGAAAU-UGGAUGCUGAUAAUUGCGCGA--GUAGGCGUGGCGUAAAAAUAAACAUCUAAG----- .................(((.((((((((.((((((....((((((.(((((....))-))).))))))...((((....--)))))))))).))))))))..)))......----- ( -32.80, z-score = -4.83, R) >droPer1.super_3 3415546 109 + 7375914 UUACAUAAACAAAUCUUUGUCAUUUUUACACCAUGCACACUCAGCAAUCAAUUGAAAU-UGGAUGCUGAUAAUUGCGCGA--GUAGGCGUGGCGUAAAAAUAAACAUCUAAA----- .................(((.((((((((.((((((....((((((.(((((....))-))).))))))...((((....--)))))))))).))))))))..)))......----- ( -32.80, z-score = -4.81, R) >droVir3.scaffold_13047 91242 101 - 19223366 --UUAAAAACAAAUUAACGUAAUUUUUACACGGCGCACAUUCAACGU------------UUGAUGCUGAAAAUAAACGAAAAACGUGCGCAGCGUAAAAAUUUGCAAUAUUU--AAC --................(((((((((((.(.((((((......(((------------(((.(......).))))))......)))))).).))))))).)))).......--... ( -23.30, z-score = -2.29, R) >droMoj3.scaffold_6540 14559364 101 + 34148556 --UAAAAAAUAAAUCAAUGUAAUUUUUACACAGCGCGCAUUUAACGUA------------UGAUGCUGAAAAUUAACGAA--ACGCGCGCGGCGUAAAAAUUGUAGAUUUUUGUAAC --......(((((....((((((((((((.(.((((((.(((..(((.------------((((.......)))))))))--).)))))).).))))))))))))....)))))... ( -25.60, z-score = -2.02, R) >droGri2.scaffold_14906 2034331 109 - 14172833 --AUAGAAGAAAUCGAAUGCAAUUUUUACACGGCGCACAUUAAACGCACAAUUGUUGGAUUGAUGCUGAGAAUUGGCGCA--AUGUGCGCAGCGUAAAAAUU--CGAUAAUU--AAC --.........(((((((.....((((((.(.(((((((((...(((.(((((.(..(.......)..).)))))))).)--)))))))).).)))))))))--))))....--... ( -35.80, z-score = -4.19, R) >consensus UUAAAUAAACAAAUCACUGUCGUUUUCACACCGUGCACACUCAACAGUCGAUUGAGU____GAUGCUGGUGAUUGC__AA__GUGUGCGAGGCGUGAAAACCAACUGCCAAC_____ .....................((((((((.(..(((((..............................................)))))..).))))))))................ (-11.60 = -10.68 + -0.92)

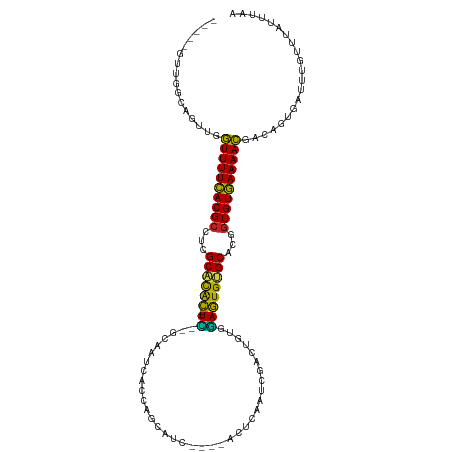
| Location | 10,728,288 – 10,728,392 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.72 |
| Shannon entropy | 0.67681 |
| G+C content | 0.42276 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -16.93 |
| Energy contribution | -15.29 |
| Covariance contribution | -1.64 |
| Combinations/Pair | 1.38 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.07 |
| SVM RNA-class probability | 0.999943 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 10728288 104 - 27905053 -----GUUGGCAGUUGGUUUUCACGCCUCGCACGC--UC--GCAGUCACCAGCAUC----ACUCAAUCGACUGUGGAGUGUGCACGGUGUGAAAACGACCGUGAUUUGUUUAUUUCA -----..(.((.((((.((((((((((..((((((--((--((((((.........----........)))))).))))))))..)))))))))))))).)).)............. ( -40.43, z-score = -2.99, R) >droSim1.chr3R 16784464 104 + 27517382 -----GUUGGCAGUUGGUUUUCACGCCUCGCACGC--UC--GCAGUCACCAGCAUC----ACUCAAUCGACUGUGGAGUGUGCACGGUGUGAAAACGACCGUGAUUUGUUUAUUUAA -----..(.((.((((.((((((((((..((((((--((--((((((.........----........)))))).))))))))..)))))))))))))).)).)............. ( -40.43, z-score = -3.18, R) >droSec1.super_0 9913591 104 + 21120651 -----GUUGGCAGUUGGUUUUCACGCCUCGCACGC--UC--GCAGUCACCAGCAUC----ACUCAAUCGACUGUGGAGUGUGCACGGUGUGAAAACGACCGUGAUUUGUUUAUUUAA -----..(.((.((((.((((((((((..((((((--((--((((((.........----........)))))).))))))))..)))))))))))))).)).)............. ( -40.43, z-score = -3.18, R) >droYak2.chr3R 13132536 104 + 28832112 -----GUUGGCAGUUGGUUUUCACGCCUCGCACAC--UU--GCAGUCACCAGCAUC----ACUCAAUCGACUGUGGAGUGUGCACGGUGUGAAAACGACGGCGAUUUGUUUAUUUAA -----....((.((((.((((((((((..((((((--(.--.(((((.........----........)))))..).))))))..)))))))))))))).))............... ( -40.33, z-score = -3.27, R) >droEre2.scaffold_4770 10264382 104 - 17746568 -----GUUGGCAGUUGGUUUUCACGCCUCGCACAC--UU--GCAGUCACCAGCAUC----ACUCAAUCGACUGUGGAGUGUGCAUGGUGUGAAAACGGCGGCGAUUUGUUUAUUUCA -----....((.((((.((((((((((..((((((--(.--.(((((.........----........)))))..).))))))..)))))))))))))).))............... ( -40.73, z-score = -3.10, R) >droAna3.scaffold_13340 5807786 106 - 23697760 -----UUAAGCUGUUAGUUUUCACGCCUCGCACAC--UUUAGCAAUCACCAGCAUC----ACUCAAUCGAUUGCAGAGUGUGCCCGAUGUGAAAACGACUGAGUUUUGUUUUUUAUA -----..((((((((.(((((((((.(..((((((--(((.((((((.........----........)))))))))))))))..).))))))))))))..)))))........... ( -32.73, z-score = -3.94, R) >dp4.chr2 20663651 109 - 30794189 -----CUUAGAUGUUUAUUUUUACGCCACGCCUAC--UCGCGCAAUUAUCAGCAUCCA-AUUUCAAUUGAUUGCUGAGUGUGCAUGGUGUAAAAAUGACAAAGAUUUGUUUAUGUAA -----......((((.((((((((((((.((....--..))(((....((((((..((-((....))))..))))))...))).))))))))))))))))................. ( -33.30, z-score = -4.06, R) >droPer1.super_3 3415546 109 - 7375914 -----UUUAGAUGUUUAUUUUUACGCCACGCCUAC--UCGCGCAAUUAUCAGCAUCCA-AUUUCAAUUGAUUGCUGAGUGUGCAUGGUGUAAAAAUGACAAAGAUUUGUUUAUGUAA -----......((((.((((((((((((.((....--..))(((....((((((..((-((....))))..))))))...))).))))))))))))))))................. ( -33.30, z-score = -4.06, R) >droVir3.scaffold_13047 91242 101 + 19223366 GUU--AAAUAUUGCAAAUUUUUACGCUGCGCACGUUUUUCGUUUAUUUUCAGCAUCAA------------ACGUUGAAUGUGCGCCGUGUAAAAAUUACGUUAAUUUGUUUUUAA-- ..(--((((...((.(((((((((((.((((((((((..(((((............))------------)))..)))))))))).)))))))))))..))..))))).......-- ( -31.50, z-score = -4.30, R) >droMoj3.scaffold_6540 14559364 101 - 34148556 GUUACAAAAAUCUACAAUUUUUACGCCGCGCGCGU--UUCGUUAAUUUUCAGCAUCA------------UACGUUAAAUGCGCGCUGUGUAAAAAUUACAUUGAUUUAUUUUUUA-- .......(((((...(((((((((((.((((((((--(((((...............------------.)))..)))))))))).))))))))))).....)))))........-- ( -29.79, z-score = -4.30, R) >droGri2.scaffold_14906 2034331 109 + 14172833 GUU--AAUUAUCG--AAUUUUUACGCUGCGCACAU--UGCGCCAAUUCUCAGCAUCAAUCCAACAAUUGUGCGUUUAAUGUGCGCCGUGUAAAAAUUGCAUUCGAUUUCUUCUAU-- ...--....((((--(((((((((((.((((((((--((((((((((.................))))).))))..))))))))).)))))))).....))))))).........-- ( -34.43, z-score = -4.91, R) >consensus _____GUUGGCAGUUGGUUUUCACGCCUCGCACAC__UC__GCAAUCACCAGCAUC____ACUCAAUCGACUGUGGAGUGUGCACGGUGUGAAAACGACAGUGAUUUGUUUAUUUAA ................((((((((((...((((((..........................................))))))...))))))))))..................... (-16.93 = -15.29 + -1.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:08 2011