
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 10,717,545 – 10,717,640 |
| Length | 95 |
| Max. P | 0.615463 |

| Location | 10,717,545 – 10,717,640 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 70.79 |
| Shannon entropy | 0.59744 |
| G+C content | 0.49554 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -14.14 |
| Energy contribution | -13.47 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.615463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 10717545 95 + 27905053 AGGACACCGGACUCUGGGGCAUUUACUUCGUGUGCGACCCUCUGCAGUGCGAGGUAAGAAGAAUUAAUGAUUCAACAGAAAACUCUCGAGAGAAA-- ...........((((.(((..(((((((((((((((......)))).)))))))))))..(((((...)))))...........))).))))...-- ( -25.00, z-score = -0.52, R) >droSim1.chr3R 16773835 95 - 27517382 AGGACACCGGACUCUGGGGCAUUUACUUCGUGUGCGACCCUCUGCAGUGCGAGGUAAGAAGAAUCAAUGAUUCGACAGAAAUCUCUCCAGAGAAA-- ...........((((((((..(((((((((((((((......)))).)))))))))))..(((((...)))))...........))))))))...-- ( -31.00, z-score = -1.70, R) >droSec1.super_0 9902979 95 - 21120651 AGGACACCGGACUCUGGGGCAUUUACUUCGUGUGCGACCCUCUGCAGUGCGAGGUAAGAAGAAUCAAUGACUCGUCAGAAAUCUCUCCAGAGAAC-- ...........((((((((..(((((((((((((((......)))).))))))))))).(((.....(((....)))....)))))))))))...-- ( -29.60, z-score = -0.94, R) >droYak2.chr3R 13121944 91 - 28832112 AGGACACCGGUCUCUGGGGCAUUUACUUCGUGUGCGACCCUCUGCAGUGUGAGGUGAGU----GCC--AGGUCUACAAUUUAGGCUUGGAAAUGGGA ......((.((.(((..(((((((((((((..((((......))).)..))))))))))----)))--(((((((.....)))))))))).)).)). ( -34.20, z-score = -1.52, R) >droEre2.scaffold_4770 10253733 91 + 17746568 AGGACACCGGUCUCUGGGGCAUUUACUUCGUCUGCGACCCUCUGCAGUGUGAGGUGAGU----GCC--AGGUCUACAAAUUAGGCUUGGAAAUGAGA ..........((((...((((((((((((..(((((......)))))...)))))))))----)))--(((((((.....)))))))......)))) ( -33.00, z-score = -1.40, R) >droAna3.scaffold_13340 13723623 93 - 23697760 AAGACACCGGUCUCUGGGGCAUUUACUUCGUGUGCGACCCUCUGCAGUGUGAGGUUGGU----ACAUAAGAAACAGAAACUACUUUCUGUUAUGUAA ......((((...)))).((((...(((.((((((.(.((((........)))).).))----))))))).((((((((....)))))))))))).. ( -26.10, z-score = -0.91, R) >dp4.chr2 7799815 84 + 30794189 AGGACACCGGUCUUUGGGGCAUUUACUUCGUGUGCGAUCCUCUGCAGUGUGAGGUAAGC----CAU--AGAUAUUCAUCCCUCCCUAAAA------- .(((.....((((....(((..((((((((..((((......))).)..))))))))))----)..--)))).....)))..........------- ( -21.80, z-score = -0.26, R) >droPer1.super_0 1623241 84 + 11822988 AGGACACCGGUCUUUGGGGCAUUUACUUCGUGUGCGAUCCUCUGCAGUGUGAGGUAAGC----CAU--AGAUAUUCAUCCCUCCCUAAAA------- .(((.....((((....(((..((((((((..((((......))).)..))))))))))----)..--)))).....)))..........------- ( -21.80, z-score = -0.26, R) >droWil1.scaffold_181130 12178734 83 - 16660200 AGGACACUGGUCUUUGGGGAAUCUAUUUCGUGUGCGAUCCCCUGCAGUGCGAGGUAAGC----UCAAAGGGAUGUCAAAA--CUAUCAA-------- ..((((....((((((((.....(((((((((((((......)))).)))))))))..)----)))))))..))))....--.......-------- ( -21.10, z-score = 0.12, R) >consensus AGGACACCGGUCUCUGGGGCAUUUACUUCGUGUGCGACCCUCUGCAGUGUGAGGUAAGA____UCA__AGAUCUACAAAAAACUCUCAAAA______ ......((((...))))....(((((((((..((((......))).)..)))))))))....................................... (-14.14 = -13.47 + -0.68)

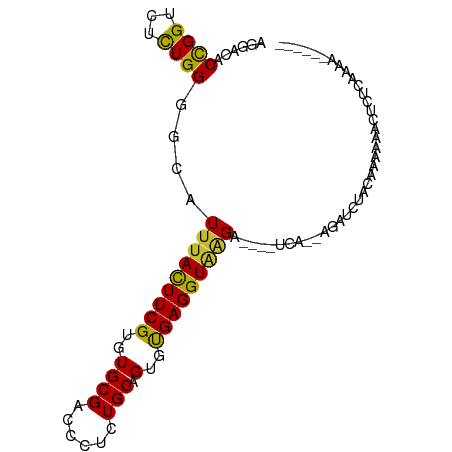
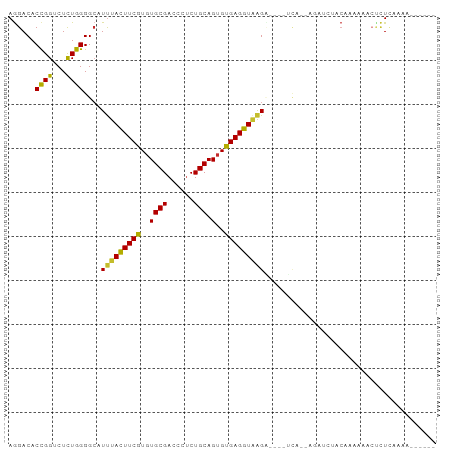
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:16:06 2011